-Search query
-Search result
Showing 1 - 50 of 218 items for (author: novacek & j)

EMDB-18065: 
Neck-tail junction of phage 812 after tail contraction (C6)
Method: single particle / : Cienikova Z, Siborova M, Fuzik T, Plevka P

EMDB-19256: 
Bacteriophage JBD30 baseplate - composite structure
Method: single particle / : Valentova L, Fuzik T, Plevka P

EMDB-19257: 
Bacteriophage JBD30 capsid - assymetric reconstruction
Method: single particle / : Valentova L, Fuzik T, Plevka P

EMDB-19258: 
Bacteriophage JBD30 procapsid computed with C5 symmetry
Method: single particle / : Valentova L, Fuzik T, Plevka P

EMDB-19259: 
Receptor binding protein of bacteriophage JBD30 computed with C3 symmetry
Method: single particle / : Valentova L, Fuzik T, Plevka P

EMDB-19260: 
Tail fibres of bacteriophage JBD30
Method: single particle / : Valentova L, Fuzik T, Plevka P

EMDB-19261: 
Baseplate of bacteriophage JBD30
Method: single particle / : Valentova L, Fuzik T, Plevka P

EMDB-19262: 
Baseplate core of bacteriophage JBD30 computed in C3 symmetry
Method: single particle / : Valentova L, Fuzik T, Plevka P

EMDB-19263: 
Baseplate of bacteriophage JBD30 computed in C3 symmetry
Method: single particle / : Valentova L, Fuzik T, Plevka P

EMDB-19264: 
Tail of bacteriophage JBD30 computed in C6 symmetry
Method: single particle / : Valentova L, Fuzik T, Plevka P

EMDB-19265: 
Stopper protein of bacteriophage JBD30 computed in C6 symmetry
Method: single particle / : Valentova L, Fuzik T, Plevka P

EMDB-19266: 
Connector complex of empty bacteriophage JBD30 particle computed in C12 symmetry
Method: single particle / : Valentova L, Fuzik T, Plevka P

EMDB-19267: 
Connector complex of bacteriophage JBD30 computed in C12 symmetry
Method: single particle / : Valentova L, Fuzik T, Plevka P

EMDB-19268: 
Capsid-connector interface of bacteriophage JBD30
Method: single particle / : Valentova L, Fuzik T, Plevka P

EMDB-19269: 
Capsid of bacteriophage JBD30 decorated with minor capsid protein trimers - asymmetric reconstruction
Method: single particle / : Valentova L, Fuzik T, Plevka P

EMDB-19270: 
Capsid of bacteriophage JBD30 decorated with minor capsid protein trimers computed in I4 symmetry
Method: single particle / : Valentova L, Fuzik T, Plevka P

EMDB-19271: 
Capsid of bacteriophage JBD30 decorated with minor capsid protein trimers computed in C5 symmetry
Method: single particle / : Valentova L, Fuzik T, Plevka P

EMDB-19272: 
Capsid of bacteriophage JBD30 - asymmetric reconstruction
Method: single particle / : Valentova L, Fuzik T, Plevka P

EMDB-19273: 
Capsid of empty bacteriophage JBD30 particle computed in C5 symmetry
Method: single particle / : Valentova L, Fuzik T, Plevka P

EMDB-19274: 
Capsid of bacteriophage JBD30 computed in C5 symmetry
Method: single particle / : Valentova L, Fuzik T, Plevka P

EMDB-19280: 
Bacteriophage JBD30 capsid
Method: single particle / : Valentova L, Fuzik T, Plevka P

EMDB-19281: 
Bacteriophage JBD30 empty particle capsid
Method: single particle / : Valentova L, Fuzik T, Plevka P

EMDB-19285: 
Procapsid of bacteriophage JBD30 computed in I4 symmetry
Method: single particle / : Valentova L, Fuzik T, Plevka P

EMDB-19439: 
Composite map of bacteriophage JBD30 capsid - neck complex
Method: single particle / : Valentova L, Fuzik T, Plevka P

EMDB-19561: 
Bacteriophage JBD30 baseplate bound to pili type IV
Method: single particle / : Valentova L, Fuzik T, Plevka P

EMDB-18048: 
Neck of phage 812 after tail contraction (C6)
Method: single particle / : Cienikova Z, Siborova M, Fuzik T, Plevka P

EMDB-17945: 
Tick-borne encephalitis virus (strain HYPR) immature particle
Method: single particle / : Fuzik T, Plevka P, Smerdova L, Novacek J

EMDB-17946: 
Tick-borne encephalitis virus (strain Neudoerfl) immature particle
Method: single particle / : Fuzik T, Plevka P, Smerdova L, Novacek J

EMDB-17947: 
Trimeric prM/E spike of Tick-borne encephalitis virus immature particle
Method: single particle / : Fuzik T, Plevka P, Smerdova L, Novacek J

PDB-8puv: 
Trimeric prM/E spike of Tick-borne encephalitis virus immature particle
Method: single particle / : Fuzik T, Plevka P, Smerdova L, Novacek J
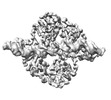
EMDB-18864: 
Central glycolytic genes regulator (CggR) bound to DNA operator
Method: single particle / : Skerlova J, Soltysova M, Rezacova P, Skubnik K

EMDB-17808: 
Tick-borne encephalitis virus Kuutsalo-14 prM3E3 trimer
Method: single particle / : Anastasina M, Domanska A, Pulkkinen LIA, Butcher SJ

EMDB-17809: 
Tick-borne encephalitis virus Kuutsalo-14 immature virus particle
Method: single particle / : Anastasina M, Domanska A, Pulkkinen LIA, Butcher SJ

EMDB-17213: 
Human Mitochondrial Lon Y186F Mutant ADP Bound
Method: single particle / : Kereiche S, Bauer JA, Matyas P, Novacek J, Kutejova E

EMDB-17214: 
Human Mitochondrial Lon Y186E Mutant ADP Bound
Method: single particle / : Kereiche S, Bauer JA, Matyas P, Novacek J, Kutejova E

PDB-8ovf: 
Human Mitochondrial Lon Y186F Mutant ADP Bound
Method: single particle / : Kereiche S, Bauer JA, Matyas P, Novacek J, Kutejova E

PDB-8ovg: 
Human Mitochondrial Lon Y186E Mutant ADP Bound
Method: single particle / : Kereiche S, Bauer JA, Matyas P, Novacek J, Kutejova E

EMDB-16923: 
Human Mitochondrial Lon Y394F Mutant ADP Bound
Method: single particle / : Kereiche S, Bauer JA, Matyas P, Novacek J, Kutejova E

EMDB-16970: 
Human Mitochondrial Lon Y186E Mutant ADP Bound
Method: single particle / : Kereiche S, Bauer JA, Matyas P, Novacek J, Kutejova E

PDB-8oka: 
Human Mitochondrial Lon Y394F Mutant ADP Bound
Method: single particle / : Kereiche S, Bauer JA, Matyas P, Novacek J, Kutejova E

PDB-8om7: 
Human Mitochondrial Lon Y186E Mutant ADP Bound
Method: single particle / : Kereiche S, Bauer JA, Matyas P, Novacek J, Kutejova E

EMDB-16915: 
Human Mitochondrial Lon Y394E Mutant ADP Bound
Method: single particle / : Kereiche S, Bauer JA, Matyas P, Novacek J, Kutejova E

PDB-8ojl: 
Human Mitochondrial Lon Y394E Mutant ADP Bound
Method: single particle / : Kereiche S, Bauer JA, Matyas P, Novacek J, Kutejova E

EMDB-19405: 
Tomograms of spoIVB Bacillus subtilis sporangia
Method: electron tomography / : Bauda E, Gallet B, Moravcova J, Effantin G, Chan H, Novacek J, Jouneau PH, Rodrigues CDA, Schoehn G, Moriscot C, Morlot C
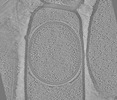
EMDB-19411: 
Tomograms of cotE Bacillus subtilis sporangia
Method: electron tomography / : Bauda E, Gallet B, Moravcova J, Effantin G, Chan H, Novacek J, Jouneau PH, Rodrigues CDA, Schoehn G, Moriscot C, Morlot C

EMDB-16546: 
structure of LEDGF/p75 PWWP domain bound to the H3K36 trimethylated dinucleosome
Method: single particle / : Koutna E, Kouba T, Novacek J, Veverka V

PDB-8cbn: 
structure of LEDGF/p75 PWWP domain bound to the H3K36 trimethylated dinucleosome
Method: single particle / : Koutna E, Kouba T, Novacek J, Veverka V

EMDB-19035: 
Composite map of the Emiliania huxleyi virus 201 (EhV-201) symmetry expanded from cryo-EM structure of virion vertex 120 nm in diameter.
Method: subtomogram averaging / : Homola M, Buttner CR, Fuzik T, Novacek J, Chaillet M, Forster F, Plevka P

EMDB-19036: 
Cryo-EM structure of the Emiliania huxleyi virus 201 (EhV-201) virion vertex with a diameter of 50 nm and a mask applied on the capsid layer.
Method: subtomogram averaging / : Homola M, Buttner CR, Fuzik T, Novacek J, Chaillet M, Forster F, Plevka P

PDB-8rbs: 
Emiliania huxleyi virus 201 (EhV-201) asymmetrical unit of capsid proteins predicted by AlphaFold2 fitted into the cryo-EM density of EhV-201 virion composite map.
Method: subtomogram averaging / : Homola M, Buttner CR, Fuzik T, Novacek J, Chaillet M, Forster F, Plevka P
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
