-Search query
-Search result
Showing 1 - 50 of 540 items for (author: ning & gao)

EMDB-37695: 
Cryo-EM structure of human SIDT1 protein with C1 symmetry at neutral pH
Method: single particle / : Liu W, Tang M, Wang J, Zhang X, Wu S, Ru H

EMDB-37696: 
Cryo-EM structure of human SIDT1 protein with C2 symmetry at neutral pH
Method: single particle / : Liu W, Tang M, Wang J, Zhang X, Wu S, Ru H

EMDB-37697: 
Cryo-EM structure of human SIDT1 protein with C1 symmetry at low pH
Method: single particle / : Liu W, Tang M, Wang J, Zhang X, Wu S, Ru H

EMDB-37698: 
Cryo-EM structure of human SIDT1 protein with C2 symmetry at low pH
Method: single particle / : Liu W, Tang M, Wang J, Zhang X, Wu S, Ru H

PDB-8woq: 
Cryo-EM structure of human SIDT1 protein with C1 symmetry at neutral pH
Method: single particle / : Liu W, Tang M, Wang J, Zhang X, Wu S, Ru H

PDB-8wor: 
Cryo-EM structure of human SIDT1 protein with C2 symmetry at neutral pH
Method: single particle / : Liu W, Tang M, Wang J, Zhang X, Wu S, Ru H

PDB-8wos: 
Cryo-EM structure of human SIDT1 protein with C1 symmetry at low pH
Method: single particle / : Liu W, Tang M, Wang J, Zhang X, Wu S, Ru H

PDB-8wot: 
Cryo-EM structure of human SIDT1 protein with C2 symmetry at low pH
Method: single particle / : Liu W, Tang M, Wang J, Zhang X, Wu S, Ru H

EMDB-38313: 
Structure of yeast replisome associated with FACT and histone hexamer, the region of FACT-Histones optimized local map
Method: single particle / : Li N, Gao Y, Yu D, Gao N, Zhai Y

EMDB-38314: 
Structure of yeast replisome associated with FACT and histone hexamer,Conformation-2
Method: single particle / : Li N, Gao Y, Yu D, Gao N, Zhai Y

EMDB-38315: 
Structure of yeast replisome associated with FACT and histone hexamer, the region of polymerase epsilon optimized local map
Method: single particle / : Li N, Gao Y, Yu D, Gao N, Zhai Y

EMDB-38316: 
Structure of yeast replisome associated with FACT and histone hexamer, Conformation-1
Method: single particle / : Li N, Gao Y, Yu D, Gao N, Zhai Y

EMDB-38317: 
Structure of yeast replisome associated with FACT and histone hexamer, Composite map
Method: single particle / : Li N, Gao Y, Yu D, Gao N, Zhai Y

PDB-8xgc: 
Structure of yeast replisome associated with FACT and histone hexamer, Composite map
Method: single particle / : Li N, Gao Y, Yu D, Gao N, Zhai Y

EMDB-35758: 
Cryo-EM structure of mouse BIRC6, N-terminal section optimized
Method: single particle / : Liu S, Jiang T, Bu F, Zhao J, Wang G, Li N, Gao N, Qiu X

EMDB-35759: 
Cryo-EM structure of mouse BIRC6, Global map
Method: single particle / : Liu S, Jiang T, Bu F, Zhao J, Wang G, Li N, Gao N, Qiu X
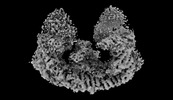
EMDB-35760: 
Cryo-EM structure of mouse BIRC6, with endogenous Smac binding
Method: single particle / : Liu S, Jiang T, Bu F, Zhao J, Wang G, Li N, Gao N, Qiu X

EMDB-38461: 
Cryo-EM structure of mouse BIRC6, Core region
Method: single particle / : Liu S, Jiang T, Bu F, Zhao J, Wang G, Li N, Gao N, Qiu X

EMDB-38462: 
Cryo-EM structure of mouse BIRC6, Half map of the core region
Method: single particle / : Liu S, Jiang T, Bu F, Zhao J, Wang G, Li N, Gao N, Qiu X

EMDB-38464: 
Cryo-EM structure of mouse BIRC6, Composite map
Method: single particle / : Liu S, Jiang T, Bu F, Zhao J, Wang G, Li N, Gao N, Qiu X

PDB-8ivq: 
Cryo-EM structure of mouse BIRC6, Global map
Method: single particle / : Liu S, Jiang T, Bu F, Zhao J, Wang G, Li N, Gao N, Qiu X
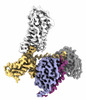
EMDB-35463: 
Cryo-EM structure of human HCAR2-Gi complex without ligand (apo state)
Method: single particle / : Pan X, Fang Y
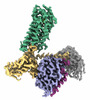
EMDB-35483: 
Cryo-EM structure of human HCAR2-Gi complex with niacin
Method: single particle / : Pan X, Fang Y
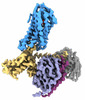
EMDB-35484: 
Cryo-EM structure of human HCAR2-Gi complex with acipimox
Method: single particle / : Pan X, Fang Y
EMDB-35485: 
Cryo-EM structure of human HCAR2-Gi complex with MK-6892
Method: single particle / : Pan X, Fang Y

PDB-8ij3: 
Cryo-EM structure of human HCAR2-Gi complex without ligand (apo state)
Method: single particle / : Pan X, Fang Y

PDB-8ija: 
Cryo-EM structure of human HCAR2-Gi complex with niacin
Method: single particle / : Pan X, Fang Y

PDB-8ijb: 
Cryo-EM structure of human HCAR2-Gi complex with acipimox
Method: single particle / : Pan X, Fang Y

PDB-8ijd: 
Cryo-EM structure of human HCAR2-Gi complex with MK-6892
Method: single particle / : Pan X, Fang Y
EMDB-37356: 
Cryo-EM structure of the GPR101-Gs complex
Method: single particle / : Sun JP, Gao N, Yu X, Wang GP, Yang F, Wang JY, Yang Z, Guan Y

EMDB-37357: 
Cryo-EM structure of the AA-14-bound GPR101-Gs complex
Method: single particle / : Sun JP, Yu X, Gao N, Yang F, Wang JY, Yang Z, Guan Y, Wang GP

EMDB-37358: 
Cryo-EM structure of the AA14-bound GPR101 complex
Method: single particle / : Sun JP, Yu X, Gao N, Yang F, Wang JY, Yang Z, Guan Y, Wang GP

PDB-8w8q: 
Cryo-EM structure of the GPR101-Gs complex
Method: single particle / : Sun JP, Gao N, Yu X, Wang GP, Yang F, Wang JY, Yang Z, Guan Y

PDB-8w8r: 
Cryo-EM structure of the AA-14-bound GPR101-Gs complex
Method: single particle / : Sun JP, Yu X, Gao N, Yang F, Wang JY, Yang Z, Guan Y, Wang GP

PDB-8w8s: 
Cryo-EM structure of the AA14-bound GPR101 complex
Method: single particle / : Sun JP, Yu X, Gao N, Yang F, Wang JY, Yang Z, Guan Y, Wang GP

EMDB-37714: 
Structure of monkeypox virus polymerase complex F8-A22-E4-H5 (tag-free A22) with exogenous DNA
Method: single particle / : Wang X, Li N, Gao N

EMDB-37715: 
Structure of monkeypox virus polymerase complex F8-A22-E4-H5 with exogenous DNA bearing one abasic site
Method: single particle / : Wang X, Li N, Gao N

EMDB-37717: 
Structure of monkeypox virus polymerase complex F8-A22-E4-H5 with exgenous DNA
Method: single particle / : Wang X, Li N, Gao N

EMDB-37722: 
Structure of monkeypox virus polymerase complex F8-A22-E4-H5 with endogenous DNA
Method: single particle / : Wang X, Li N, Gao N

PDB-8wpe: 
Structure of monkeypox virus polymerase complex F8-A22-E4-H5 (tag-free A22) with exogenous DNA
Method: single particle / : Wang X, Li N, Gao N

PDB-8wpf: 
Structure of monkeypox virus polymerase complex F8-A22-E4-H5 with exogenous DNA bearing one abasic site
Method: single particle / : Wang X, Li N, Gao N

PDB-8wpk: 
Structure of monkeypox virus polymerase complex F8-A22-E4-H5 with exgenous DNA
Method: single particle / : Wang X, Li N, Gao N

PDB-8wpp: 
Structure of monkeypox virus polymerase complex F8-A22-E4-H5 with endogenous DNA
Method: single particle / : Wang X, Li N, Gao N

EMDB-34724: 
Cryo-EM structure of CpcL-PBS from cyanobacterium Synechocystis sp. PCC 6803
Method: single particle / : Zheng L, Zhang Z, Wang H, Zheng Z, Gao N, Zhao J

PDB-8hfq: 
Cryo-EM structure of CpcL-PBS from cyanobacterium Synechocystis sp. PCC 6803
Method: single particle / : Zheng L, Zhang Z, Wang H, Zheng Z, Gao N, Zhao J

EMDB-33174: 
Structure of nucleosome-AAG complex (T-50I, post-catalytic state)
Method: single particle / : Zheng L, Tsai B, Gao N

PDB-7xfj: 
Structure of nucleosome-AAG complex (T-50I, post-catalytic state)
Method: single particle / : Zheng L, Tsai B, Gao N

EMDB-34948: 
Cryo-EM structure of the apo-GPR132-Gi
Method: single particle / : Wang JL, Ding JH, Sun JP, Yu X

EMDB-34950: 
Activation mechanism of GPR132 by NPGLY
Method: single particle / : Wang JL, Ding JH, Sun JP, Yu X

EMDB-34951: 
Activation mechanism of GPR132 by 9(S)-HODE
Method: single particle / : Wang JL, Ding JH, Sun JP, Yu X
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
