-Search query
-Search result
Showing 1 - 50 of 209 items for (author: murakami & k)

EMDB-42502: 
Cyanobacterial RNA polymerase elongation complex with NusG and CTP
Method: single particle / : Qayyum MZ, Imashimizu M, Leanca M, Vishwakarma RK, Bradley Riaz A, Yuzenkova Y, Murakami KS

PDB-8urw: 
Cyanobacterial RNA polymerase elongation complex with NusG and CTP
Method: single particle / : Qayyum MZ, Imashimizu M, Leanca M, Vishwakarma RK, Bradley Riaz A, Yuzenkova Y, Murakami KS

EMDB-40874: 
Cyanobacterial RNAP-EC
Method: single particle / : Qayyum MZ, Imashimizu M, Leanca M, Vishwakarma RK, Bradley Riaz A, Yuzenkova Y, Murakami KS

PDB-8syi: 
Cyanobacterial RNAP-EC
Method: single particle / : Qayyum MZ, Imashimizu M, Leanca M, Vishwakarma RK, Bradley Riaz A, Yuzenkova Y, Murakami KS

EMDB-27500: 
EBNA1 DNA binding domain (DBD) (458-617)+2 repeats of family repeat (FR) region
Method: single particle / : Mei Y, Lieberman PM, Murakami K

PDB-8dlf: 
EBNA1 DNA binding domain (DBD) (458-617)+2 repeats of family repeat (FR) region
Method: single particle / : Mei Y, Lieberman PM, Murakami K

EMDB-28162: 
SARS-CoV-2 polyprotein substrate regulates the stepwise Mpro cleavage reaction
Method: single particle / : Narwal M, Edwards T, Armache JP, Murakami KS

EMDB-28200: 
Cryo-EM structure of SARS CoV-2 Mpro WT protease
Method: single particle / : Narwal M, Edwards T, Armache JP, Murakami KS

PDB-8eir: 
SARS-CoV-2 polyprotein substrate regulates the stepwise Mpro cleavage reaction
Method: single particle / : Narwal M, Edwards T, Armache JP, Murakami KS

PDB-8eke: 
Cryo-EM structure of SARS CoV-2 Mpro WT protease
Method: single particle / : Narwal M, Edwards T, Armache JP, Murakami KS

EMDB-26547: 
Mediator-PIC Early (Tail A + Upstream DNA & Activator)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

EMDB-26551: 
Mediator-PIC Early (Composite Model)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

PDB-7uik: 
Mediator-PIC Early (Tail A + Upstream DNA & Activator)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

PDB-7uio: 
Mediator-PIC Early (Composite Model)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

EMDB-26542: 
Core Mediator-PICearly (Copy A)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

EMDB-26543: 
Mediator-PIC Early (Tail A)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

EMDB-26544: 
Mediator-PIC Early (Core B)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

EMDB-26545: 
Mediator-PIC Early (Mediator A)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K
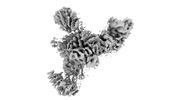
EMDB-26548: 
Mediator-PIC Early (Tail A/B Dimer)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

EMDB-28023: 
Dimerized Mediator-PIC with early GTFs and TFIIE on a divergent promoter
Method: subtomogram averaging / : Palao III L
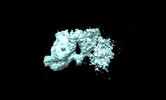
EMDB-28024: 
Dimerized Mediator PIC with early GTFs on a divergent promoter
Method: subtomogram averaging / : Palao III L

PDB-7ui9: 
Core Mediator-PICearly (Copy A)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

PDB-7uic: 
Mediator-PIC Early (Tail A)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

PDB-7uif: 
Mediator-PIC Early (Core B)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

PDB-7uig: 
Mediator-PIC Early (Mediator A)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

PDB-7uil: 
Mediator-PIC Early (Tail A/B Dimer)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

EMDB-28149: 
Mycobacterium tuberculosis paused transcription complex with Bacillus subtilis NusG
Method: single particle / : Vishwakarma RK, Murakami KS

EMDB-28174: 
M. tuberculosis RNAP pause escaped complex with Bacillus subtilis NusG and GMPCPP
Method: single particle / : Vishwakarma RK, Murakami KS

EMDB-28373: 
Mycobacterium tuberculosis transcription elongation complex with Bacillus subtilis NusG (EC_LG)
Method: single particle / : Vishwakarm RK, Murakami KS

EMDB-28374: 
Mycobacterium tuberculosis transcription elongation complex with Bacillus subtilis NusG (EC_PG)
Method: single particle / : Vishwakarm RK, Murakami KS

EMDB-28466: 
M. tuberculosis RNAP elongation complex with NusG and CMPCPP
Method: single particle / : Vishwakarma RK, Murakami KS

EMDB-28467: 
M. tuberculosis RNAP elongation complex with NusG
Method: single particle / : Vishwakarma RK, Murakami KS

EMDB-28665: 
M. tuberculosis RNAP paused complex with B. subtilis NusG and GMPCPP
Method: single particle / : Vishwakarma RK, Murakami KS

PDB-8ehq: 
Mycobacterium tuberculosis paused transcription complex with Bacillus subtilis NusG
Method: single particle / : Vishwakarma RK, Murakami KS

PDB-8ej3: 
M. tuberculosis RNAP pause escaped complex with Bacillus subtilis NusG and GMPCPP
Method: single particle / : Vishwakarma RK, Murakami KS

PDB-8eoe: 
Mycobacterium tuberculosis transcription elongation complex with Bacillus subtilis NusG (EC_LG)
Method: single particle / : Vishwakarm RK, Murakami KS

PDB-8eof: 
Mycobacterium tuberculosis transcription elongation complex with Bacillus subtilis NusG (EC_PG)
Method: single particle / : Vishwakarm RK, Murakami KS

PDB-8eos: 
M. tuberculosis RNAP elongation complex with NusG and CMPCPP
Method: single particle / : Vishwakarma RK, Murakami KS

PDB-8eot: 
M. tuberculosis RNAP elongation complex with NusG
Method: single particle / : Vishwakarma RK, Murakami KS

PDB-8exy: 
M. tuberculosis RNAP paused complex with B. subtilis NusG and GMPCPP
Method: single particle / : Vishwakarma RK, Murakami KS

EMDB-26481: 
Cryo-EM Structure of Bl_Man38A nucleophile mutant in complex with mannose at 2.7 A
Method: single particle / : Santos CR, Cordeiro RL, Domingues MN, Borges AC, de Farias MA, Van Heel M, Murakami MT, Portugal RV

PDB-7ufu: 
Cryo-EM Structure of Bl_Man38A nucleophile mutant in complex with mannose at 2.7 A
Method: single particle / : Santos CR, Cordeiro RL, Domingues MN, Borges AC, de Farias MA, Van Heel M, Murakami MT, Portugal RV

EMDB-26480: 
Cryo-EM Structure of Bl_Man38C at 2.9 A
Method: single particle / : Santos CR, Cordeiro RL, Domingues MN, Borges AC, de Farias MA, Van Heel M, Murakami MT, Portugal RV

PDB-7uft: 
Cryo-EM Structure of Bl_Man38C at 2.9 A
Method: single particle / : Santos CR, Cordeiro RL, Domingues MN, Borges AC, de Farias MA, Van Heel M, Murakami MT, Portugal RV

EMDB-26479: 
Cryo-EM Structure of Bl_Man38B at 3.4 A
Method: single particle / : Santos CR, Cordeiro RL, Domingues MN, Borges AC, de Farias MA, Van Heel M, Murakami MT, Portugal RV

PDB-7ufs: 
Cryo-EM Structure of Bl_Man38B at 3.4 A
Method: single particle / : Santos CR, Cordeiro RL, Domingues MN, Borges AC, de Farias MA, Van Heel M, Murakami MT, Portugal RV

EMDB-26478: 
Cryo-EM Structure of Bl_Man38A at 2.7 A
Method: single particle / : Santos CR, Cordeiro RL, Domingues MN, Borges AC, de Farias MA, Van Heel M, Murakami MT, Portugal RV

PDB-7ufr: 
Cryo-EM Structure of Bl_Man38A at 2.7 A
Method: single particle / : Santos CR, Cordeiro RL, Domingues MN, Borges AC, de Farias MA, Van Heel M, Murakami MT, Portugal RV

EMDB-27989: 
Octameric prenyltransferase domain of fusicoccadiene Synthase with C2 symmetry sans transiently associating cyclase domains
Method: single particle / : Faylo JL, van Eeuwen T, Christianson DW

EMDB-28046: 
Symmetry-expanded consensus map of PaFS prenyltransferase core with single interacting cyclase domain
Method: single particle / : Faylo JL, van Eeuwen T, Christianson DW
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
