-Search query
-Search result
Showing 1 - 50 of 161 items for (author: mukherjee & s)

EMDB-19440: 
Cryo-EM structure of human NTCP-Bulevirtide complex
Method: single particle / : Liu H, Zakrzewicz D, Nosol K, Irobalieva RN, Mukherjee S, Bang-Soerensen R, Goldmann N, Kunz S, Rossi L, Kossiakoff AA, Urban S, Glebe D, Geyer J, Locher KP

PDB-8rqf: 
Cryo-EM structure of human NTCP-Bulevirtide complex
Method: single particle / : Liu H, Zakrzewicz D, Nosol K, Irobalieva RN, Mukherjee S, Bang-Soerensen R, Goldmann N, Kunz S, Rossi L, Kossiakoff AA, Urban S, Glebe D, Geyer J, Locher KP

EMDB-18520: 
Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes
Method: single particle / : Moura TR, Purta E, Bernat A, Baulin E, Mukherjee S, Bujnicki JM

EMDB-18521: 
Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes
Method: single particle / : Moura TR, Purta E, Bernat A, Baulin E, Mukherjee S, Bujnicki JM

EMDB-18522: 
Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes
Method: single particle / : Moura TR, Purta E, Bernat A, Baulin E, Mukherjee S, Bujnicki JM

EMDB-18523: 
Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes
Method: single particle / : Moura TR, Purta E, Bernat A, Baulin E, Mukherjee S, Bujnicki JM

PDB-8qo2: 
Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes
Method: single particle / : Moura TR, Purta E, Bernat A, Baulin E, Mukherjee S, Bujnicki JM
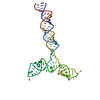
PDB-8qo3: 
Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes
Method: single particle / : Moura TR, Purta E, Bernat A, Baulin E, Mukherjee S, Bujnicki JM

PDB-8qo4: 
Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes
Method: single particle / : Moura TR, Purta E, Bernat A, Baulin E, Mukherjee S, Bujnicki JM

PDB-8qo5: 
Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes
Method: single particle / : Moura TR, Purta E, Bernat A, Baulin E, Mukherjee S, Bujnicki JM
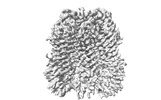
EMDB-34266: 
CryoEM structure of human Pannexin1 with R217H congenital mutation.
Method: single particle / : Hussain N, Penmatsa A

EMDB-34267: 
Cryo-EM structure of human Pannexin1 resembling Pannexin2 pore with W74R/R75Dmutations
Method: single particle / : Hussain N, Penmatsa A

EMDB-34265: 
CryoEM structure of human Pannexin isoform 3
Method: single particle / : Hussain N, Penmatsa A
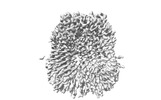
EMDB-34268: 
human Pannexin1
Method: single particle / : Hussain N, Penmatsa A, Vinothkumar KR

EMDB-43033: 
Structure of 80alpha portal protein expressed in E. coli
Method: single particle / : Kizziah JL, Mukherjee A, Dokland T

EMDB-43142: 
SaPI1 mature capsid structure containing DNA
Method: single particle / : Mukherjee A, Kizziah JL, Dokland T

EMDB-43143: 
SaPI1 mature capsid structure without DNA
Method: single particle / : Mukherjee A, Kizziah JL, Dokland T

EMDB-43145: 
SaPI1 portal structure in mature capsids containing DNA
Method: single particle / : Kizziah JL, Mukherjee A, Dokland T

EMDB-43146: 
SaPI1 portal structure in mature capsids without DNA
Method: single particle / : Mukherjee A, Kizziah JL, Dokland T

EMDB-43147: 
SaPI1 portal-capsid interface in mature capsids with DNA
Method: single particle / : Kizziah JL, Mukherjee A, Dokland T

PDB-8v8b: 
Structure of 80alpha portal protein expressed in E. coli
Method: single particle / : Kizziah JL, Mukherjee A, Dokland T

PDB-8vd4: 
SaPI1 mature capsid structure containing DNA
Method: single particle / : Mukherjee A, Kizziah JL, Dokland T

PDB-8vd5: 
SaPI1 mature capsid structure without DNA
Method: single particle / : Mukherjee A, Kizziah JL, Dokland T

PDB-8vd8: 
SaPI1 portal structure in mature capsids containing DNA
Method: single particle / : Kizziah JL, Mukherjee A, Dokland T

PDB-8vdc: 
SaPI1 portal structure in mature capsids without DNA
Method: single particle / : Mukherjee A, Kizziah JL, Dokland T

PDB-8vde: 
SaPI1 portal-capsid interface in mature capsids with DNA
Method: single particle / : Kizziah JL, Mukherjee A, Dokland T

EMDB-17655: 
Human OATP1B3
Method: single particle / : Ciuta AD, Nosol K, Kowal J, Mukherjee S, Ramirez AS, Stieger B, Kossiakoff AA, Locher KP

EMDB-17677: 
Human OATP1B1
Method: single particle / : Ciuta AD, Nosol K, Kowal J, Mukherjee S, Ramirez AS, Stieger B, Kossiakoff AA, Locher KP

PDB-8pg0: 
Human OATP1B3
Method: single particle / : Ciuta AD, Nosol K, Kowal J, Mukherjee S, Ramirez AS, Stieger B, Kossiakoff AA, Locher KP

PDB-8phw: 
Human OATP1B1
Method: single particle / : Ciuta AD, Nosol K, Kowal J, Mukherjee S, Ramirez AS, Stieger B, Kossiakoff AA, Locher KP
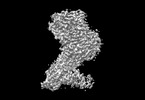
EMDB-36294: 
Legionella effector protein SidI
Method: single particle / : Wang L, Subramanian A, Mukherjee S, Walter P

PDB-8jhu: 
Legionella effector protein SidI
Method: single particle / : Wang L, Subramanian A, Mukherjee S, Walter P
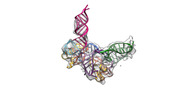
EMDB-34047: 
Cryo-EM structure of Pseudomonas aeruginosa RsmZ RNA in complex with two RsmA protein dimers
Method: single particle / : Jia X, Pan Z, Yuan Y, Luo B, Luo Y, Mukherjee S, Jia G, Ling X, Yang X, Wu Y, Liu T, Wei X, Bujnick JM, Zhao K, Su Z
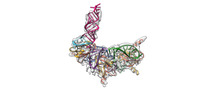
EMDB-34048: 
Cryo-EM structure of Pseudomonas aeruginosa RsmZ RNA in complex with three RsmA protein dimers
Method: single particle / : Jia X, Pan Z, Yuan Y, Luo B, Luo Y, Mukherjee S, Jia G, Liu L, Ling X, Yang X, Wu Y, Liu T, Miao Z, Wei X, Bujnicki JM, Zhao K, Su Z

PDB-7yr6: 
Cryo-EM structure of Pseudomonas aeruginosa RsmZ RNA in complex with two RsmA protein dimers
Method: single particle / : Jia X, Pan Z, Yuan Y, Luo B, Luo Y, Mukherjee S, Jia G, Ling X, Yang X, Wu Y, Liu T, Wei X, Bujnick JM, Zhao K, Su Z

PDB-7yr7: 
Cryo-EM structure of Pseudomonas aeruginosa RsmZ RNA in complex with three RsmA protein dimers
Method: single particle / : Jia X, Pan Z, Yuan Y, Luo B, Luo Y, Mukherjee S, Jia G, Liu L, Ling X, Yang X, Wu Y, Liu T, Miao Z, Wei X, Bujnicki JM, Zhao K, Su Z
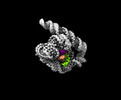
EMDB-33136: 
The Tet-S2 state of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z
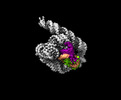
EMDB-33137: 
The Tet-S2 state with a pseudoknotted 4-way junction of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z

PDB-7xd5: 
The Tet-S2 state of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z

PDB-7xd6: 
The Tet-S2 state with a pseudoknotted 4-way junction of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z
EMDB-33134: 
The relaxed pre-Tet-S1 state of wild-type Tetrahymena group I intron with 6nt 3'/5'-exon
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z

EMDB-33135: 
The intermediate pre-Tet-S1 state of wild-type Tetrahymena group I intron with 6nt 3'/5'-exon
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z

EMDB-33138: 
The pre-Tet-C state of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z
EMDB-34670: 
The relaxed pre-Tet-S1 state of G264A mutated Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z
EMDB-34671: 
The intermediate pre-Tet-S1 state of G264A mutated Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z
EMDB-35223: 
The Tet-S1 state of G264A mutated Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z

PDB-7xd3: 
The relaxed pre-Tet-S1 state of wild-type Tetrahymena group I intron with 6nt 3'/5'-exon
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z

PDB-7xd4: 
The intermediate pre-Tet-S1 state of wild-type Tetrahymena group I intron with 6nt 3'/5'-exon
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z

PDB-7xd7: 
The pre-Tet-C state of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z

PDB-8hd6: 
The relaxed pre-Tet-S1 state of G264A mutated Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
