-Search query
-Search result
Showing 1 - 50 of 122 items for (author: moore & mj)

EMDB-41888: 
Structure of Apo CXCR4/Gi complex
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC

EMDB-41889: 
Structure of CXCL12-bound CXCR4/Gi complex
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC

EMDB-41890: 
Structure of AMD3100-bound CXCR4/Gi complex
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC

EMDB-41891: 
Structure of REGN7663 Fab-bound CXCR4/Gi complex
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC

EMDB-41892: 
Structure of REGN7663-Fab bound CXCR4
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC

EMDB-41893: 
Structure of trimeric CXCR4 in complex with REGN7663 Fab
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC

EMDB-41894: 
Structure of tetrameric CXCR4 in complex with REGN7663 Fab
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC

EMDB-42124: 
Cryo-EM structure of human STEAP1 in complex with AMG 509 Fab
Method: single particle / : Li F, Bailis JM

EMDB-28570: 
CryoEM structure of PN45545 TCR-CD3 complex
Method: single particle / : Saotome K, Franklin MC

EMDB-28571: 
CryoEM structure of PN45545 TCR-CD3 in complex with HLA-A2 MAGEA4 (230-239)
Method: single particle / : Saotome K, Franklin MC

EMDB-28572: 
CryoEM structure of PN45428 TCR-CD3 in complex with HLA-A2 MAGEA4
Method: single particle / : Saotome K, Franklin MC

EMDB-28573: 
CryoEM structure of HLA-A2 bound to MAGEA4 (230-239) peptide
Method: single particle / : Saotome K, Franklin MC

EMDB-28574: 
CryoEM structure of HLA-A2 bound to MAGEA8 (232-241) peptide
Method: single particle / : Saotome K, Franklin MC

EMDB-28080: 
Human cardiac myosin II and associated essential light chain in the rigor conformation
Method: single particle / : Doran MH, Lehman W, Rynkiewicz MJ

EMDB-28081: 
Human beta-cardiac myosin II bound to ADP-MG2+ and the associated essential light chain
Method: single particle / : Doran MH, Lehman W, Rynkiewicz MJ

EMDB-28082: 
Helical reconstruction of the human cardiac actin-tropomyosin-myosin complex in complex with ADP-Mg2+
Method: helical / : Doran MH, Lehman W, Rynkiewicz MJ

EMDB-28083: 
Helical reconstruction of the human cardiac actin-tropomyosin-myosin complex in the rigor form
Method: helical / : Doran MH, Lehman W, Rynkiewicz MJ

EMDB-28270: 
Helical reconstruction of the human cardiac actin-tropomyosin-myosin loop 4 7G mutant complex
Method: helical / : Doran MH, Lehman W, Rynkiewicz MJ

EMDB-25810: 
Human Metapneumovirus F trimer in complex with M4B06 Fab
Method: single particle / : Xiao X, Tian D, Yan X, Eddins NJ, Galli JD, Su H, Vora KA, Chen Z, Zhang L

EMDB-24675: 
AMC018 SOSIP.v4.2 in complex with PGV04 Fab
Method: single particle / : Cottrell CA, de Val N, Ward AB

EMDB-24676: 
AMC016 SOSIP.v4.2 in complex with PGV04 Fab
Method: single particle / : Bader DLV, Cottrell CA, Ward AB
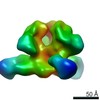
EMDB-24243: 
RM12K in complex with BG505 SOSIP
Method: single particle / : Cottrell CA, Ward AB

EMDB-24244: 
RM54B1 Fab in complex with BG505 SOSIP
Method: single particle / : Cottrell CA, Ward AB

EMDB-24245: 
RM15E Fab in complex with BG505 SOSIP
Method: single particle / : Cottrell CA, Ward AB
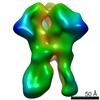
EMDB-24246: 
RM15A Fab in complex with BG505 SOSIP
Method: single particle / : Cottrell CA, Ward AB
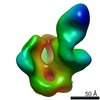
EMDB-24247: 
RM35A1 Fab in complex with BG505 SOSIP
Method: single particle / : Cottrell CA, Ward AB

EMDB-22170: 
BG505 SOSIPv5.2 in complex with PGT122 and two RM20E1 Fabs
Method: single particle / : Cottrell CA, Ward AB

EMDB-22171: 
BG505 SOSIPv5.2 in complex with PGT122 and three RM20E1 Fabs
Method: single particle / : Cottrell CA, Ward AB

EMDB-22172: 
C3 symmetric BG505 SOSIPv5.2 in complex with PGT122 and three RM20E1 Fabs
Method: single particle / : Cottrell CA, Ward AB

EMDB-22173: 
BG505 SOSIPv5.2 in complex with PGT122 and one RM20E1 Fab
Method: single particle / : Cottrell CA, Ward AB

EMDB-22178: 
BG505 SOSIPv5.2 in complex with PGT122 and no RM20E1 Fabs
Method: single particle / : Cottrell CA, Ward AB

EMDB-22179: 
C3 symmetric BG505 SOSIPv5.2 in complex with PGT122 and no RM20E1 Fabs
Method: single particle / : Cottrell CA, Ward AB

EMDB-22714: 
nsEM map of the 16055 SOSIP HIV Env trimer in complex with polyclonal Fab from rabbit r2463
Method: single particle / : Ward AB, Antanasijevic A

EMDB-22715: 
nsEM map of the 16055 SOSIP HIV Env trimer in complex with polyclonal Fab from rabbit r2464
Method: single particle / : Ward AB, Antanasijevic A

EMDB-22716: 
nsEM map of the 16055 SOSIP HIV Env trimer in complex with polyclonal Fab from rabbit r2465
Method: single particle / : Ward AB, Antanasijevic A

EMDB-22717: 
nsEM map of the 16055 SOSIP HIV Env trimer in complex with polyclonal Fab from rabbit r2466
Method: single particle / : Ward AB, Antanasijevic A

EMDB-22718: 
nsEM map of the 16055 SOSIP HIV Env trimer in complex with polyclonal Fab from rabbit r2467
Method: single particle / : Ward AB, Antanasijevic A

EMDB-22719: 
nsEM map of the 16055 SOSIP HIV Env trimer in complex with polyclonal Fab from rabbit r2468
Method: single particle / : Ward AB, Antanasijevic A

EMDB-22720: 
nsEM map of the 16055 SOSIP HIV Env trimer in complex with polyclonal Fab from rabbit r2469
Method: single particle / : Ward AB, Antanasijevic A

EMDB-22721: 
nsEM map of the 16055 SOSIP HIV Env trimer in complex with polyclonal Fab from rabbit r2470
Method: single particle / : Ward AB, Antanasijevic A

EMDB-22722: 
nsEM map of the 16055 SOSIP HIV Env trimer in complex with polyclonal Fab from rabbit r2471
Method: single particle / : Ward AB, Antanasijevic A

EMDB-22723: 
nsEM map of the 16055 SOSIP HIV Env trimer in complex with polyclonal Fab from rabbit r2472
Method: single particle / : Ward AB, Antanasijevic A

EMDB-21258: 
AMC009 SOSIP.v4.2 in complex with PGV04 Fab
Method: single particle / : Cottrell CA, de Val N, Ward AB
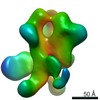
EMDB-22434: 
UA0068 week 38 Fabs in complex with REJO SOSIP.v4.2 D368R
Method: single particle / : Cottrell CA, Torres JL, Sewall LM, Ward AB
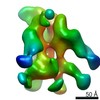
EMDB-22435: 
UA0088 week 38 Fabs in complex with SF162p3 SOSIP.v9
Method: single particle / : Cottrell CA, Torres JL, Sewall LM, Ward AB
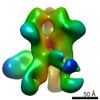
EMDB-22436: 
UA0083 week 38 Fabs in complex with SF162p3 SOSIP.v9
Method: single particle / : Cottrell CA, Torres JL, Sewall LM, Ward AB
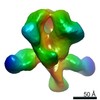
EMDB-22437: 
UA0089 week 38 Fabs in complex with AMCO11 SOSIP.v5.2
Method: single particle / : Cottrell CA, Torres JL, Sewall LM, Ward AB
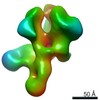
EMDB-22438: 
UA0089 week 38 Fabs in complex with AMCO11 SOSIP.v5.2
Method: single particle / : Cottrell CA, Torres JL, Sewall LM, Ward AB
EMDB-22439: 
UA0091 week 38 Fabs in complex with REJO SOSIP.v4.2 D368R
Method: single particle / : Cottrell CA, Torres JL, Sewall LM, Ward AB
EMDB-22440: 
UA0091 week 38 Fabs in complex with REJO SOSIP.v4.2 D368R
Method: single particle / : Cottrell CA, Torres JL, Sewall LM, Ward AB
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
