-Search query
-Search result
Showing 1 - 50 of 120 items for (author: montoya & g)

EMDB-15698: 
Cryo-EM structure of Cas12k-sgRNA binary complex (type V-K CRISPR-associated transposon)
Method: single particle / : Tenjo-Castano F, Sofos N, Stella S, Molina R, Pape T, Lopez-Mendez B, Stutzke LS, Temperini P, Montoya G

PDB-8axb: 
Cryo-EM structure of Cas12k-sgRNA binary complex (type V-K CRISPR-associated transposon)
Method: single particle / : Tenjo-Castano F, Sofos N, Stella S, Molina R, Pape T, Lopez-Mendez B, Stutzke LS, Temperini P, Montoya G

EMDB-15697: 
Cryo-EM structure of shCas12k-sgRNA-dsDNA ternary complex (type V-K CRISPR-associated transposon)
Method: single particle / : Tenjo-Castano F, Sofos N, Stella S, Fuglsang A, Pape T, Mesa P, Stutzke LS, Temperini P, Montoya G

PDB-8axa: 
Cryo-EM structure of shCas12k-sgRNA-dsDNA ternary complex (type V-K CRISPR-associated transposon)
Method: single particle / : Tenjo-Castano F, Sofos N, Stella S, Fuglsang A, Pape T, Mesa P, Stutzke LS, Temperini P, Montoya G

EMDB-28163: 
Cryo-EM map of octopus sensory receptor CRT1
Method: single particle / : Kang G, Kim JJ, Allard CAH, Valencia-Montoya WA, Bellono NW, Hibbs RE

EMDB-28167: 
Cryo-EM map of squid sensory receptor CRB1
Method: single particle / : Kang G, Kim JJ, Allard CAH, Valencia-Montoya WA, van Giesen L, Kilian PB, Bai X, Bellono NW, Hibbs RE

PDB-8eis: 
Cryo-EM structure of octopus sensory receptor CRT1
Method: single particle / : Kang G, Kim JJ, Allard CAH, Valencia-Montoya WA, Bellono NW, Hibbs RE

PDB-8eiz: 
Cryo-EM structure of squid sensory receptor CRB1
Method: single particle / : Kang G, Kim JJ, Allard CAH, Valencia-Montoya WA, van Giesen L, Kilian PB, Bai X, Bellono NW, Hibbs RE

EMDB-16016: 
ISDra2 TnpB in complex with reRNA
Method: single particle / : Sasnauskas G, Tamulaitiene G, Carabias A, Siksnys V, Montoya G, Druteika G, Silanskas A, Venclovas C, Karvelis T, Kazlauskas D

PDB-8bf8: 
ISDra2 TnpB in complex with reRNA
Method: single particle / : Sasnauskas G, Tamulaitiene G, Carabias A, Siksnys V, Montoya G, Druteika G, Silanskas A, Venclovas C, Karvelis T, Kazlauskas D
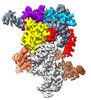
EMDB-28655: 
ISDra2 TnpB in complex with reRNA and cognate DNA, conformation 2 (RuvC domain unresolved)
Method: single particle / : Sasnauskas G, Tamulaitiene G, Carabias A, Karvelis T, Druteika G, Silanskas A, Montoya G, Venclovas C, Kazlauskas D, Siksnys V

EMDB-28656: 
ISDra2 TnpB in complex with reRNA and cognate DNA, conformation 1 (RuvC domain resolved)
Method: single particle / : Sasnauskas G, Tamulaitiene G, Carabias A, Karvelis T, Druteika G, Silanskas A, Montoya G, Venclovas C, Kazlauskas D, Siksnys V

PDB-8ex9: 
ISDra2 TnpB in complex with reRNA and cognate DNA, conformation 2 (RuvC domain unresolved)
Method: single particle / : Sasnauskas G, Tamulaitiene G, Carabias A, Karvelis T, Druteika G, Silanskas A, Montoya G, Venclovas C, Kazlauskas D, Siksnys V

PDB-8exa: 
ISDra2 TnpB in complex with reRNA and cognate DNA, conformation 1 (RuvC domain resolved)
Method: single particle / : Sasnauskas G, Tamulaitiene G, Carabias A, Karvelis T, Druteika G, Silanskas A, Montoya G, Venclovas C, Kazlauskas D, Siksnys V

EMDB-25419: 
Previously uncharacterized rectangular bacteria in the dolphin mouth
Method: electron tomography / : Dudek NK, Galaz-Montoya JG, Shi H, Mayer M, Danita C, Celis AI, Wu GH, Behr B, Huang KC, Chiu W, Relman DA
EMDB-29207: 
CryoET tomogram of mitochondria in BACHD mouse model neuron
Method: electron tomography / : Wu GH, Galaz-Montoya JG, Gu Y, Mitchell PG, Wu C, Chiu W
EMDB-29208: 
CryoET tomogram of BACHD mouse model neuron showing sheet aggregates
Method: electron tomography / : Wu GH, Galaz-Montoya JG, Gu Y, Mitchell PG, Wu C, Chiu W
EMDB-29210: 
CryoET tomogram of purified mitochondria from HD patient iPSC-derived neuron (Q109)
Method: electron tomography / : Wu GH, Smith-Geater C, Galaz-Montoya JG, Mitchell PG, Thompson LM, Chiu W
EMDB-29211: 
CryoET tomogram of HD patient iPSC-derived neuron (Q66) with PIAS1 hetKO treatment
Method: electron tomography / : Wu GH, Smith-Geater C, Galaz-Montoya JG, Mitchell PG, Thompson LM, Chiu W

EMDB-28668: 
CryoET tomogram of iPSC-derived control non-HD neuron (Q18)
Method: electron tomography / : Wu GH, Smith-Geater C, Galaz-Montoya JG, Mitchell PG, Thompson LM, Chiu W
EMDB-28944: 
CryoET tomogram of iPSC-derived control non-HD neuron (Q20)
Method: electron tomography / : Wu GH, Smith-Geater C, Galaz-Montoya JG, Mitchell PG, Thompson LM, Chiu W
EMDB-28946: 
CryoET tomogram of HD patient iPSC-derived neuron (Q53)
Method: electron tomography / : Wu GH, Smith-Geater C, Galaz-Montoya JG, Mitchell PG, Thompson LM, Chiu W
EMDB-29074: 
CryoET tomogram of HD patient iPSC-derived neuron (Q66)
Method: electron tomography / : Wu GH, Smith-Geater C, Galaz-Montoya JG, Mitchell PG, Thompson LM, Chiu W

EMDB-29075: 
CryoET tomogram of HD patient iPSC-derived neuron (Q77)
Method: electron tomography / : Wu GH, Smith-Geater C, Galaz-Montoya JG, Mitchell PG, Thompson LM, Chiu W

EMDB-29076: 
CryoET tomogram of HD patient iPSC-derived neuron (Q109)
Method: electron tomography / : Wu GH, Smith-Geater C, Galaz-Montoya JG, Mitchell PG, Thompson LM, Chiu W

EMDB-29079: 
CryoET tomogram of HD patient iPSC-derived neuron (Q66) showing sheet aggregate
Method: electron tomography / : Wu GH, Smith-Geater C, Galaz-Montoya JG, Mitchell PG, Thompson LM, Chiu W

EMDB-29080: 
CryoET tomogram of HD patient iPSC-derived neuron (Q66) with PIAS1 hetKO treatment
Method: electron tomography / : Wu GH, Smith-Geater C, Galaz-Montoya JG, Mitchell PG, Thompson LM, Chiu W
EMDB-29081: 
CryoET tomogram of HD patient iPSC-derived neuron (Q66) with GRSF1 KD treatment
Method: electron tomography / : Wu GH, Smith-Geater C, Galaz-Montoya JG, Mitchell PG, Thompson LM, Chiu W

EMDB-29083: 
CryoET tomogram of mitochondria in WT mouse neuron
Method: electron tomography / : Wu GH, Galaz-Montoya JG, Gu Y, Mitchell PG, Wu C, Chiu W

EMDB-29084: 
CryoET tomogram of mitochondria in BACHD-dN17 mouse model neuron
Method: electron tomography / : Wu GH, Galaz-Montoya JG, Gu Y, Mitchell PG, Wu C, Chiu W

EMDB-15294: 
Cryo-EM structure of the strand transfer complex of the TnsB transposase (type V-K CRISPR-associated transposon)
Method: single particle / : Tenjo-Castano F, Sofos N, Lopez-Mendez B, Stutzke LS, Fuglsang A, Stella S, Montoya G

EMDB-15344: 
Type V-K CAST TnsB bound to LTR-SR
Method: single particle / : Tenjo-Castano F, Sofos N, Lopez-Mendez B, Stutzke LS, Fuglsang A, Stella S, Montoya G

PDB-8aa5: 
Cryo-EM structure of the strand transfer complex of the TnsB transposase (type V-K CRISPR-associated transposon)
Method: single particle / : Tenjo-Castano F, Sofos N, Lopez-Mendez B, Stutzke LS, Fuglsang A, Stella S, Montoya G

EMDB-14472: 
Structure of the RAF1-HSP90-CDC37 complex (RHC-II)
Method: single particle / : Mesa P, Garcia-Alonso S, Barbacid M, Montoya G

EMDB-14473: 
Structure of the RAF1-HSP90-CDC37 complex (RHC-I)
Method: single particle / : Mesa P, Garcia-Alonso S, Barbacid M, Montoya G

PDB-7z37: 
Structure of the RAF1-HSP90-CDC37 complex (RHC-II)
Method: single particle / : Mesa P, Garcia-Alonso S, Barbacid M, Montoya G

PDB-7z38: 
Structure of the RAF1-HSP90-CDC37 complex (RHC-I)
Method: single particle / : Mesa P, Garcia-Alonso S, Barbacid M, Montoya G

EMDB-13619: 
Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ATPgS, state III (composite map)
Method: single particle / : Saleh A, Noguchi Y, Aramayo R, Ivanova ME, Speck C

EMDB-13620: 
Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ADP:BeF3, state I
Method: single particle / : Saleh A, Noguchi Y, Aramayo R, Ivanova ME, Speck C

EMDB-13621: 
Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ATPgS, state III (B1 map)
Method: single particle / : Saleh A, Noguchi Y, Aramayo R, Ivanova ME, Speck C

EMDB-13622: 
Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ATPgS, state III (B2 map)
Method: single particle / : Saleh A, Noguchi Y, Aramayo R, Ivanova ME, Speck C

EMDB-13623: 
Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ATPgS, state III (B3 map)
Method: single particle / : Saleh A, Noguchi Y, Aramayo R, Ivanova ME, Speck C

EMDB-13624: 
Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ATPgS, state III (3D auto-refined map)
Method: single particle / : Saleh A, Noguchi Y, Aramayo R, Ivanova ME, Speck C

EMDB-13629: 
Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ATPgS, state II (composite map)
Method: single particle / : Saleh A, Noguchi Y, Aramayo R, Ivanova ME, Speck C

EMDB-13631: 
Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ATPgS, state II (B1 map)
Method: single particle / : Saleh A, Noguchi Y, Aramayo R, Ivanova ME, Speck C

EMDB-13635: 
Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ATPgS, state II (B2 map)
Method: single particle / : Saleh A, Noguchi Y, Aramayo R, Ivanova ME, Speck C

EMDB-13640: 
Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ATPgS, state II (3D auto-refined map)
Method: single particle / : Saleh A, Noguchi Y, Aramayo R, Ivanova ME, Speck C

EMDB-13644: 
Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ATPgS, state I (3D auto-refined map)
Method: single particle / : Saleh A, Noguchi Y, Aramayo R, Ivanova ME, Speck C

EMDB-13645: 
Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ADP:BeF3, state I (B1 map)
Method: single particle / : Saleh A, Noguchi Y, Aramayo R, Ivanova ME, Speck C

EMDB-13646: 
Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ADP:BeF3, state I (B2 map)
Method: single particle / : Saleh A, Noguchi Y, Aramayo R, Ivanova ME, Speck C
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
