-Search query
-Search result
Showing 1 - 50 of 237 items for (author: montefiori & dc)
EMDB-40242: 
BG505 MD39 SOSIP in complex with Rh.NJ85 wk12 gp120GH, N611/FP, and base epitope polyclonal antibodies
Method: single particle / : Lee WH, Ozorowski G, Torres JL, Ward AB

EMDB-40243: 
BG505 MD39 SOSIP in complex with Rh.NJ86 wk12 V1V3, C3V5, N611/FP, and base epitope polyclonal antibodies
Method: single particle / : Lee WH, Ozorowski G, Torres JL, Ward AB

EMDB-40244: 
BG505 MD39 SOSIP in complex with Rh.NJ76 wk12 C3V5, N611/FP, and base epitope polyclonal antibodies
Method: single particle / : Lee WH, Ozorowski G, Torres JL, Ward AB
EMDB-40252: 
BG505 MD39 SOSIP in complex with Rh.NK04 wk12 gp120 glycan hole and base epitope polyclonal antibodies
Method: single particle / : Lee WS, Ozorowski G, Torres JL, Ward AB

EMDB-40254: 
BG505 MD39 SOSIP in complex with Rh.NJ75 wk12 gp120 glycan hole and base epitope polyclonal antibodies
Method: single particle / : Lee WS, Ozorowski G, Torres JL, Ward AB

EMDB-40255: 
BG505 MD39 SOSIP in complex with Rh.NJ87 wk12 C3V5 and base epitope polyclonal antibodies
Method: single particle / : Lee WS, Ozorowski G, Torres JL, Ward AB

EMDB-40256: 
BG505 MD39 SOSIP in complex with Rh.NJ84 wk12 V1V3, gp120 glycan hole and base epitope polyclonal antibodies
Method: single particle / : Lee WS, Ozorowski G, Torres JL, Ward AB

EMDB-40257: 
BG505 MD39 SOSIP in complex with Rh.NJ77 wk12 V1V3, C3V5, N611/FP and base epitope polyclonal
Method: single particle / : Lee WS, Ozorowski G, Torres JL, Ward AB

EMDB-27703: 
Structure of RBD directed antibody DH1047 in complex with SARS-CoV-2 spike: Local refinement of RBD-Fab interace
Method: single particle / : May AJ, Manne K, Acharya P

EMDB-14888: 
CM01B in complex with ConM SOSIP
Method: single particle / : van Schooten J, Ward AB

EMDB-14889: 
CM05A1 in complex with conM SOSIP
Method: single particle / : van Schooten J, Ward AB

EMDB-14890: 
CM02A in complex with conM SOSIP
Method: single particle / : van Schooten J, Ward AB

EMDB-14891: 
CM04F in complex with conM SOSIP
Method: single particle / : van Schooten J, Ward AB

EMDB-14892: 
CM03B in complex with conM SOSIP
Method: single particle / : van Schooten J, Ward AB

EMDB-14893: 
CM05E in complex with conM SOSIP
Method: single particle / : van Schooten J, Ward AB

EMDB-14894: 
CM05L in complex with conM SOSIP
Method: single particle / : van Schooten J, Ward AB

EMDB-14895: 
NHP MCM02 wk26 N355/N289 pAb in complex with ConM SOSIP
Method: single particle / : van Schooten J, Ward AB

EMDB-14896: 
NHP MCM04 wk26 base pAb in complex with ConM SOSIP
Method: single particle / : van Schooten J, Ward AB

EMDB-14897: 
NHP MCM04 wk26 N611 pAb in complex with ConM SOSIP
Method: single particle / : van Schooten J, Ward AB

EMDB-14898: 
NHP MCM05 wk26 V1V2V3 and base pAbs in complex with ConM SOSIP
Method: single particle / : van Schooten J, Ward AB

EMDB-14899: 
NHP MCM06 wk26 base and N611 pAbs in complex with ConS SOSIP
Method: single particle / : van Schooten J, Ward AB

EMDB-14900: 
NHP MCM02 wk26 N355/N289 pAb in complex with ConS SOSIP
Method: single particle / : van Schooten J, Ward AB

EMDB-14901: 
NHP CCK046 wk70 base pAb in complex with ConM SOSIP
Method: single particle / : van Schooten J, Ward AB

EMDB-14902: 
NHP MCM06 wk70 N355/N289 pAb in complex with ConM SOSIP
Method: single particle / : van Schooten J, Ward AB

EMDB-14903: 
NHP MCM06 wk70 N611 pAb in complex with ConM SOSIP
Method: single particle / : van Schooten J, Ward AB

EMDB-14904: 
NHP MCM04 wk70 V1V2V3 pAb in complex with ConM SOSIP
Method: single particle / : van Schooten J, Ward AB

EMDB-14905: 
NHP MCM03 wk70 base pAb in complex with ConS SOSIP
Method: single particle / : van Schooten J, Ward AB

EMDB-14906: 
NHP MCM04 wk70 N611 pAb in complex with ConS SOSIP
Method: single particle / : van Schooten J, Ward AB

EMDB-14907: 
NHP MCM04 wk70 V1V2V3 pAb in complex with ConS SOSIP
Method: single particle / : van Schooten J, Ward AB

EMDB-14908: 
NHP MCM06 wk70 N355/N289 pAb in complex with ConS SOSIP
Method: single particle / : van Schooten J, Ward AB

EMDB-14784: 
AMC009 SOSIPv5.2 + ACS110 Fab
Method: single particle / : van Schooten J, Ward A

EMDB-14785: 
AMC009 SOSIPv.52 in complex with ACS114 Fab
Method: single particle / : van Schooten J, Ward A

EMDB-14786: 
AMC009 SOSIPv5.2 in complex with ACS117 Fab
Method: single particle / : van Schooten J, Ward A

EMDB-14787: 
AMC009 SOSIPv5.2 in complex with ACS122 Fab
Method: single particle / : van Schooten J, Ward A

EMDB-14788: 
AMC009 SOSIPv5.2 in complex with ACS125 Fab
Method: single particle / : van Schooten J, Ward A

EMDB-14789: 
AMC009 SOSIPv5.2 in complex with ACS131 Fab
Method: single particle / : van Schooten J, Ward A

EMDB-27610: 
BG505 MD39 SOSIP in complex with Rh.NJ82 wk13 N611 and base epitope pAbs
Method: single particle / : Torres JL, Lee WH, Ozorowski G, Ward AB
EMDB-27611: 
BG505 MD39 SOSIP in complex with Rh.NJ95 wk13 base epitope pAb
Method: single particle / : Torres JL, Lee WH, Ozorowski G, Ward AB
EMDB-27612: 
BG505 MD39 SOSIP in complex with Rh.NK05 wk13 base epitope pAb
Method: single particle / : Torres JL, Lee WH, Ozorowski G, Ward AB
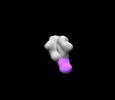
EMDB-27614: 
BG505 MD39 SOSIP in complex with Rh.NJ79 wk13 base epitope pAb
Method: single particle / : Torres JL, Lee WH, Ozorowski G, Ward AB

EMDB-27615: 
BG505 MD39 SOSIP in complex with Rh.NJ93 wk13 base and V5/C3 epitope pAbs
Method: single particle / : Torres JL, Lee WH, Ozorowski G, Ward AB

EMDB-14783: 
AMC009 SOSIPv5.2 in complex with Fabs ACS101 and ACS124
Method: single particle / : van Schooten J, Ozorowski G, Ward A

EMDB-27601: 
BG505 MD39 SOSIP in complex with Rh.K409 wk13 base, FP/gp41, V5/C3 and N611 epitope pAbs
Method: single particle / : Sewall LM, Richey ST, Ozorowski G, Ward AB

EMDB-27602: 
BG505 MD39 SOSIP in complex with Rh.K487 wk13 base, FP/gp41, and V5/C3 epitope pAbs
Method: single particle / : Sewall LM, Richey ST, Ozorowski G, Ward AB
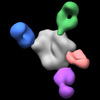
EMDB-27603: 
BG505 MD39 SOSIP in complex with Rh.DGT5 wk13 base, N335/N289, V1/V3 and V5/C3 epitope pAbs
Method: single particle / : Sewall LM, Richey ST, Ozorowski G, Ward AB
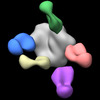
EMDB-27604: 
BG505 MD39 SOSIP in complex with Rh.K949 wk33 base, N335/N289, V1/V3, N611 and V5/C3 epitope pAbs
Method: single particle / : Sewall LM, Richey ST, Ozorowski G, Ward AB

EMDB-27605: 
BG505 MD39 SOSIP in complex with Rh.L282 wk33 base, N335/N289, FP/gp41 and V5/C3 epitope pAbs
Method: single particle / : Sewall LM, Richey ST, Ozorowski G, Ward AB

EMDB-27607: 
BG505 MD39 SOSIP in complex with Rh.DHHT wk33 base, FP/gp41 and V5/C3 epitope pAbs
Method: single particle / : Sewall LM, Richey ST, Ozorowski G, Ward AB

EMDB-27608: 
BG505 MD39 SOSIP in complex with Rh.DHID wk33 base, N355/N289 and V1/V3 epitope pAbs
Method: single particle / : Sewall LM, Richey ST, Ozorowski G, Ward AB

EMDB-27609: 
BG505 MD39 SOSIP in complex with Rh.NJ78 wk13 V5/C3 and base epitope pAbs
Method: single particle / : Torres JL, Lee WH, Ozorowski G, Ward AB
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
