-Search query
-Search result
Showing all 48 items for (author: miyata & t)

EMDB-36190: 
Cryo-EM Structure of Na-dithionite Reduced Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus
Method: single particle / : Suzuki Y, Miyata T, Makino F, Tanaka H, Namba K, Sowa K, Kitazumi Y, Shirai O

EMDB-36191: 
Cryo-EM Structure of K-ferricyanide Oxidized Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus
Method: single particle / : Suzuki Y, Miyata T, Makino F, Tanaka H, Namba K, Sowa K, Kitazumi Y, Shirai O

PDB-8jej: 
Cryo-EM Structure of Na-dithionite Reduced Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus
Method: single particle / : Suzuki Y, Miyata T, Makino F, Tanaka H, Namba K, Sowa K, Kitazumi Y, Shirai O

PDB-8jek: 
Cryo-EM Structure of K-ferricyanide Oxidized Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus
Method: single particle / : Suzuki Y, Miyata T, Makino F, Tanaka H, Namba K, Sowa K, Kitazumi Y, Shirai O

EMDB-34871: 
Cryo-EM structure of biparatopic antibody Bp109-92 in complex with TNFR2
Method: single particle / : Akiba H, Fujita J, Ise T, Nishiyama K, Miyata T, Kato T, Namba K, Ohno H, Kamada H, Nagata S, Tsumoto K

PDB-8hlb: 
Cryo-EM structure of biparatopic antibody Bp109-92 in complex with TNFR2
Method: single particle / : Akiba H, Fujita J, Ise T, Nishiyama K, Miyata T, Kato T, Namba K, Ohno H, Kamada H, Nagata S, Tsumoto K

EMDB-34368: 
Cryo-EM Structure of Membrane-Bound Alcohol Dehydrogenase from Gluconobacter oxydans
Method: single particle / : Adachi T, Miyata T, Makino F, Tanaka H, Namba K, Sowa K, Kitazumi Y, Shirai O

EMDB-34369: 
Cryo-EM Structure of Membrane-Bound Aldehyde Dehydrogenase from Gluconobacter oxydans
Method: single particle / : Adachi T, Miyata T, Makino F, Tanaka H, Namba K, Sowa K, Kitazumi Y, Shirai O

PDB-8gy2: 
Cryo-EM Structure of Membrane-Bound Alcohol Dehydrogenase from Gluconobacter oxydans
Method: single particle / : Adachi T, Miyata T, Makino F, Tanaka H, Namba K, Sowa K, Kitazumi Y, Shirai O

PDB-8gy3: 
Cryo-EM Structure of Membrane-Bound Aldehyde Dehydrogenase from Gluconobacter oxydans
Method: single particle / : Adachi T, Miyata T, Makino F, Tanaka H, Namba K, Sowa K, Kitazumi Y, Shirai O

EMDB-32764: 
Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus
Method: single particle / : Suzuki Y, Makino F

PDB-7wsq: 
Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus
Method: single particle / : Suzuki Y, Makino F, Miyata T, Tanaka H, Namba K, Sowa K, Kitazumi Y, Shirai O

EMDB-32262: 
Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus
Method: single particle / : Suzuki Y, Makino F, Miyata T, Tanaka H, Namba K, Sowa K, Kitazumi Y, Shirai O

PDB-7w2j: 
Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus
Method: single particle / : Suzuki Y, Makino F, Miyata T, Tanaka H, Namba K, Sowa K, Kitazumi Y, Shirai O

EMDB-31944: 
Pentacylindrical allophycocyanin core from Thermosynechococcus vulcanus
Method: single particle / : Kawakami K, Hamaguchi T, Hirose Y, Kosumi D, Miyata M, Kamiya N, Yonekura K

PDB-7vea: 
Pentacylindrical allophycocyanin core from Thermosynechococcus vulcanus
Method: single particle / : Kawakami K, Hamaguchi T, Hirose Y, Kosumi D, Miyata M, Kamiya N, Yonekura K

EMDB-31945: 
Phycocyanin rod structure of cyanobacterial phycobilisome
Method: single particle / : Kawakami K, Hamaguchi T, Hirose Y, Kosumi D, Miyata M, Kamiya N, Yonekura K

PDB-7veb: 
Phycocyanin rod structure of cyanobacterial phycobilisome
Method: single particle / : Kawakami K, Hamaguchi T, Hirose Y, Kosumi D, Miyata M, Kamiya N, Yonekura K

EMDB-32151: 
Cryo-EM Structure of Formate Dehydrogenase 1 from Methylorubrum extorquens AM1
Method: single particle / : Yoshikawa T, Makino F, Miyata T, Suzuki Y, Tanaka H, Namba K, Sowa K, Kitazumi Y, Shirai O

PDB-7vw6: 
Cryo-EM Structure of Formate Dehydrogenase 1 from Methylorubrum extorquens AM1
Method: single particle / : Yoshikawa T, Makino F, Miyata T, Suzuki Y, Tanaka H, Namba K, Sowa K, Kitazumi Y, Shirai O

EMDB-31520: 
Negative stain map of chained F1-like ATPase complex of Mycoplasma mobile
Method: single particle / : Toyonaga T, Miyata M

EMDB-30398: 
CryoEM structure of S.typhimurium flagellar LP ring
Method: single particle / : Yamaguchi T, Makino F, Miyata T, Minamino T, Kato T, Namba K

EMDB-30409: 
CryoEM structure of S.typhimurium flagellar hook-basal body (HBB) complex
Method: single particle / : Yamaguchi T, Makino F, Miyata T, Minamino T, Kato T, Namba K

PDB-7clr: 
CryoEM structure of S.typhimurium flagellar LP ring
Method: single particle / : Yamaguchi T, Makino F, Miyata T, Minamino T, Kato T, Namba K

EMDB-30360: 
34-fold symmetry salmonella MS ring
Method: single particle / : Kawamoto A, Miyata T, Makino F, Kinoshita M, Minamino T, Imada K, Kato T, Namba K

EMDB-30361: 
11 fold-syymetry of salmonella MS ring
Method: single particle / : Kawamoto A, Miyata T, Makino F, Kinoshita M, Minamino T, Imada K, Kato T, Namba K

EMDB-30363: 
Without imposing symmetry of salmonella MS ring
Method: single particle / : Kawamoto A, Miyata T, Makino F, Kinoshita M, Minamino T, Imada K, Kato T, Namba K

EMDB-30613: 
Structure of the bacterial flagellar basal body
Method: single particle / : Kawamoto A, Miyata T, Makino F, Kinoshita M, Minamino T, Imada K, Kato T, Namba K

EMDB-30940: 
Salmonella flagella MS-ring protein FliF 1-503 having C33 symmetry
Method: single particle / : Makino F, Kawamoto A, Namba K

EMDB-30941: 
Salmonella flagella MS-ring protein FliF 1-456 having C35 symmetry
Method: single particle / : Makino F, Kawamoto A, Namba K

EMDB-30942: 
Salmonella flagella MS-ring protein FliF 1-456 having C34 symmetry
Method: single particle / : Makino F, Kawamoto A, Namba K
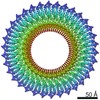
EMDB-30612: 
34-fold symmetry Salmonella S ring formed by full-length FliF
Method: single particle / : Kawamoto A, Miyata T

PDB-7d84: 
34-fold symmetry Salmonella S ring formed by full-length FliF
Method: single particle / : Kawamoto A, Miyata T, Makino F, Kinoshita M, Minamino T, Imada K, Kato T, Namba K

EMDB-30233: 
Cryo-EM structure of the human pathogen Mycoplasma pneumoniae P1
Method: single particle / : Kawamoto A, Kenri T

PDB-7bwm: 
Cryo-EM structure of the human pathogen Mycoplasma pneumoniae P1
Method: single particle / : Kawamoto A, Kenri T, Namba K, Miyata M

EMDB-0980: 
CryoEM structure of S.typhimurium R-type straight flagellar filament made of FljB (A461V)
Method: single particle / : Yamaguchi T, Toma S

EMDB-9896: 
CryoEM structure of S.typhimurium R-type flagellar filament made of FljB (A461V) without domain D3 by masking out
Method: helical / : Yamaguchi T, Toma S

PDB-6jy0: 
CryoEM structure of S.typhimurium R-type straight flagellar filament made of FljB (A461V)
Method: helical / : Yamaguchi T, Toma S, Terahara N, Miyata T, Minamino T, Ashikara M, Namba K, Kato T

EMDB-9952: 
Structure of the native supercoiled hook as a universal joint
Method: single particle / : Kato T, Miyata T

PDB-6k9q: 
Structure of the native supercoiled hook as a universal joint
Method: single particle / : Kato T, Miyata T, Makino F, Horvath P, Namba K

EMDB-0893: 
Detailed structures of the gliding machinery observed by ECT
Method: electron tomography / : Kawamoto A, Kato T, Namba K

EMDB-9974: 
Structure of Salmonella flagellar hook reveals intermolecular domain interactions for the universal joint function
Method: helical / : Horvath P, Kato T

PDB-6kfk: 
Structure of Salmonella flagellar hook reveals intermolecular domain interactions for the universal joint function
Method: helical / : Horvath P, Kato T, Miyata T, Namba K

EMDB-6683: 
The 7 angstrom resolution CryoEM map of the bacterial flagellar polyrod
Method: helical / : Fujii T, Namba K

PDB-5wrh: 
FlgG structure based on the CryoEM map of the bacterial flagellar polyrod
Method: helical / : Fujii T, Namba K

EMDB-2520: 
Injectisome in the tomogram of Salmonella mini-cell
Method: subtomogram averaging / : Kawamoto A, Morimoto VY, Miyata T, Minamino T, Hughes TK, Kato T, Namba K
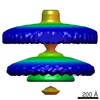
EMDB-2521: 
Flagellar hook basal body in the tomogram of Salmonella mini-cell
Method: subtomogram averaging / : Kawamoto A, Morimoto VY, Miyata T, Minamino T, Hughes TK, Kato T, Namba K
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
