-Search query
-Search result
Showing all 21 items for (author: milne & jls)

EMDB-7770: 
Atomic resolution cryo-EM structure of beta-galactosidase
Method: single particle / : Subramaniam S, Bartesaghi A, Banerjee S, Zhu X

EMDB-6630: 
Glutamate dehydrogenase in the unliganded state
Method: single particle / : Borgnia MJ, Banerjee S, Merk A, Matthies D, Bartesaghi A, Rao P, Pierson J, Earl LA, Falconieri V, Subramaniam S, Milne JLS

EMDB-6631: 
Glutamate dehydrogenase in complex with GTP
Method: single particle / : Borgnia MJ, Banerjee S, Merk A, Matthies D, Bartesaghi A, Rao P, Pierson J, Earl LA, Falconieri V, Subramaniam S, Milne JLS

EMDB-6632: 
Glutamate dehydrogenase in complex with NADH and GTP, open conformation
Method: single particle / : Borgnia MJ, Banerjee S, Merk A, Matthies D, Bartesaghi A, Rao P, Pierson J, Earl LA, Falconieri V, Subramaniam S, Milne JLS

EMDB-6633: 
Glutamate dehydrogenase in complex with NADH and GTP, closed conformation
Method: single particle / : Borgnia MJ, Banerjee S, Merk A, Matthies D, Bartesaghi A, Rao P, Pierson J, Earl LA, Falconieri V, Subramaniam S, Milne JLS

EMDB-6634: 
Glutamate dehydrogenase in complex with NADH, closed conformation
Method: single particle / : Borgnia MJ, Banerjee S, Merk A, Matthies D, Bartesaghi A, Rao P, Pierson J, Earl LA, Falconieri V, Subramaniam S, Milne JLS

EMDB-6635: 
Glutamate dehydrogenase in complex with NADH, open conformation
Method: single particle / : Borgnia MJ, Banerjee S, Merk A, Matthies D, Bartesaghi A, Rao P, Pierson J, Earl LA, Falconieri V, Subramaniam S, Milne JLS

EMDB-3295: 
Cryo-EM structure of human p97 bound UPCDC30245 inhibitor
Method: single particle / : Banerjee S, Bartesaghi A, Merk A, Rao P, Bulfer SL, Yan Y, Green N, Mroczkowski B, Neitz RJ, Wipf P, Falconieri V, Deshaies RJ, Milne JLS, Huryn D, Arkin M, Subramaniam S

EMDB-3296: 
Cryo-EM structure of human p97 bound to ADP
Method: single particle / : Banerjee S, Bartesaghi A, Merk A, Rao P, Bulfer SL, Yan Y, Green N, Mroczkowski B, Neitz RJ, Wipf P, Falconieri V, Deshaies RJ, Milne JLS, Huryn D, Arkin M, Subramaniam S

EMDB-3297: 
Cryo-EM structure of human p97 bound to ATPgS (Conformation I)
Method: single particle / : Banerjee S, Bartesaghi A, Merk A, Rao P, Bulfer SL, Yan Y, Green N, Mroczkowski B, Neitz RJ, Wipf P, Falconieri V, Deshaies RJ, Milne JLS, Huryn D, Arkin M, Subramaniam S

EMDB-3298: 
Cryo-EM structure of human p97 bound to ATPgS (Conformation II)
Method: single particle / : Banerjee S, Bartesaghi A, Merk A, Rao P, Bulfer SL, Yan Y, Green N, Mroczkowski B, Neitz RJ, Wipf P, Falconieri V, Deshaies RJ, Milne JLS, Huryn D, Arkin M, Subramaniam S

EMDB-3299: 
Cryo-EM structure of human p97 bound to ATPgS (Conformation III)
Method: single particle / : Banerjee S, Bartesaghi A, Merk A, Rao P, Bulfer SL, Yan Y, Green N, Mroczkowski B, Neitz RJ, Wipf P, Falconieri V, Deshaies RJ, Milne JLS, Huryn D, Arkin M, Subramaniam S

EMDB-2484: 
Pre-fusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy
Method: single particle / : Bartesaghi A, Merk A, Borgnia MJ, Milne JLS, Subramaniam S

EMDB-5455: 
Cryo-electron tomography of native, trimeric HIV-1 BaL Env in complex with soluble 2-domain CD4
Method: subtomogram averaging / : Tran EEH, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JLS, Subramaniam S

EMDB-5456: 
Cryo-electron tomography of native, trimeric HIV-1 BaL Env in complex with 17b IgG
Method: subtomogram averaging / : Tran EEH, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JLS, Subramaniam S

EMDB-5457: 
Cryo-electron tomography of native, trimeric HIV-1 BaL Env in complex with VRC01 IgG
Method: subtomogram averaging / : Tran EEH, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JLS, Subramaniam S

EMDB-5458: 
Cryo-electron tomography of native, trimeric HIV-1 BaL Env in complex with VRC03 IgG
Method: subtomogram averaging / : Tran EEH, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JLS, Subramaniam S
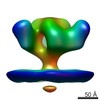
EMDB-5459: 
Cryo-electron tomography of native, trimeric HIV-1 BaL Env in complex with VRC02 Fab
Method: subtomogram averaging / : Tran EEH, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JLS, Subramaniam S

EMDB-5460: 
Cryo-electron tomography of native, trimeric HIV-1 BaL Env in complex with VRC02 IgG
Method: subtomogram averaging / : Tran EEH, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JLS, Subramaniam S
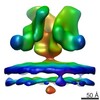
EMDB-5461: 
Cryo-electron tomography of native, trimeric HIV-1 BaL Env in complex with VRC01 IgG and 17b IgG
Method: subtomogram averaging / : Tran EEH, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JLS, Subramaniam S

EMDB-5462: 
Cryo-electron tomography of a trimeric soluble HIV Env construct, gp140 SOSIP, in complex with Fab 17b
Method: subtomogram averaging / : Tran EEH, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JLS, Subramaniam S
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
