-Search query
-Search result
Showing all 33 items for (author: michael & saur)

EMDB-17208: 
CRYO-EM STRUCTURE OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A

EMDB-17212: 
CRYO-EM STRUCTURE OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : TB11CS6H1 snoRNA mutant
Method: single particle / : Rajan KS, Yonath A

EMDB-17249: 
CRYO-EM CONSENSUS MAP OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A
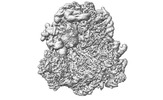
EMDB-17250: 
CRYO-EM FOCUSED REFINEMENT MAPS OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A

EMDB-17254: 
CRYO-EM CONSENSUS MAP OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : TB11CS6H1 SKO
Method: single particle / : RAJAN KS, YONATH A
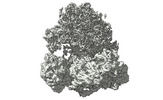
EMDB-17255: 
CRYO-EM FOCUSED REFINEMENT OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : TB11CS6H1 snoRNA mutant
Method: single particle / : Rajan KS, Yonath A

PDB-8ova: 
CRYO-EM STRUCTURE OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A

PDB-8ove: 
CRYO-EM STRUCTURE OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : TB11CS6H1 snoRNA mutant
Method: single particle / : Rajan KS, Yonath A

EMDB-10574: 
Beta-galactosidase in complex with PETG
Method: single particle / : Saur M, Hartshorn MJ, Dong J, Reeks J, Bunkoczi G, Jhoti H, Williams PA

EMDB-10575: 
PKM2 in complex with Compound 5
Method: single particle / : Saur M, Hartshorn MJ, Dong J, Reeks J, Bunkoczi G, Jhoti H, Williams PA

EMDB-10576: 
PKM2 in complex with L-threonine
Method: single particle / : Saur M, Hartshorn MJ, Dong J, Reeks J, Bunkoczi G, Jhoti H, Williams PA

EMDB-10577: 
PKM2 in complex with Compound 6
Method: single particle / : Saur M, Hartshorn MJ, Dong J, Reeks J, Bunkoczi G, Jhoti H, Williams PA

EMDB-10584: 
PKM2 in complex with Compound 10
Method: single particle / : Saur M, Hartshorn MJ, Dong J, Reeks J, Bunkoczi G, Jhoti H, Williams PA

PDB-6tte: 
Beta-galactosidase in complex with PETG
Method: single particle / : Saur M, Hartshorn MJ, Dong J, Reeks J, Bunkoczi G, Jhoti H, Williams PA

PDB-6ttf: 
PKM2 in complex with Compound 5
Method: single particle / : Saur M, Hartshorn MJ, Dong J, Reeks J, Bunkoczi G, Jhoti H, Williams PA

PDB-6tth: 
PKM2 in complex with L-threonine
Method: single particle / : Saur M, Hartshorn MJ, Dong J, Reeks J, Bunkoczi G, Jhoti H, Williams PA

PDB-6tti: 
PKM2 in complex with Compound 6
Method: single particle / : Saur M, Hartshorn MJ, Dong J, Reeks J, Bunkoczi G, Jhoti H, Williams PA

PDB-6ttq: 
PKM2 in complex with Compound 10
Method: single particle / : Saur M, Hartshorn MJ, Dong J, Reeks J, Bunkoczi G, Jhoti H, Williams PA

EMDB-10563: 
Beta-galactosidase in complex with deoxygalacto-nojirimycin
Method: single particle / : Saur M, Hartshorn MJ, Dong J, Reeks J, Bunkoczi G, Jhoti H, Williams PA

EMDB-10564: 
Beta-galactosidase in complex with L-ribose
Method: single particle / : Saur M, Hartshorn MJ, Dong J, Reeks J, Bunkoczi G, Jhoti H, Williams PA

PDB-6tsh: 
Beta-galactosidase in complex with deoxygalacto-nojirimycin
Method: single particle / : Saur M, Hartshorn MJ, Dong J, Reeks J, Bunkoczi G, Jhoti H, Williams PA

PDB-6tsk: 
Beta-galactosidase in complex with L-ribose
Method: single particle / : Saur M, Hartshorn MJ, Dong J, Reeks J, Bunkoczi G, Jhoti H, Williams PA

EMDB-3717: 
Structure of differently sized plant protein
Method: single particle / : Saur M, Hennig R, Young P, Rusitzka K, Hellmann N, Heidrich J, Morgner N, Markl J, Schneider D

EMDB-3737: 
Structure of IM30 in C10-symmetrical assembly
Method: single particle / : Saur M, Hennig R, Young P, Rusitzka K, Hellmann N, Heidrich J, Morgner N, Markl J, Schneider D

EMDB-3738: 
Structure of IM30 in C12-symmetrical assembly
Method: single particle / : Saur M, Hennig R, Young P, Rusitzka K, Hellmann N, Heidrich J, Morgner N, Markl J, Schneider D

EMDB-3739: 
Structure of IM30 in C13-symmetrical assembly
Method: single particle / : Saur M, Hennig R, Young P, Rusitzka K, Hellmann N, Heidrich J, Morgner N, Markl J, Schneider D

EMDB-3740: 
Structure of IM30 in C14-symmetrical assembly
Method: single particle / : Saur M, Hennig R, Young P, Rusitzka K, Hellmann N, Heidrich J, Morgner N, Markl J, Schneider D

EMDB-3645: 
CryoEM density of TcdA1 in prepore state (SPHIRE tutorial)
Method: single particle / : Raunser S, Gatsogiannis S, Roderer S

EMDB-4103: 
Structure of the human Rod-Zw10-Zwilch (RZZ) complex
Method: single particle / : Mosalaganti S, Keller J

EMDB-4104: 
Negative stain reconstruction of the ROD(1-1250):Zwilch:ZW10
Method: single particle / : Mosalaganti S

EMDB-2055: 
Acetylcholine-binding protein in the hemolymph of the planorbid snail Biomphalaria glabrata is a pentagonal dodecahedron (60 subunits)
Method: single particle / : Saur M, Moeller V, Kapetanopoulos K, Braukmann S, Gebauer W, Tenzer S, Markl J

PDB-4aod: 
Biomphalaria glabrata Acetylcholine-binding protein type 1 (BgAChBP1)
Method: single particle / : Saur M, Moeller V, Kapetanopoulos K, Braukmann S, Gebauer W, Tenzer S, Markl J

PDB-4aoe: 
Biomphalaria glabrata Acetylcholine-binding protein type 2 (BgAChBP2)
Method: single particle / : Saur M, Moeller V, Kapetanopoulos K, Braukmann S, Gebauer W, Tenzer S, Markl J
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
