-Search query
-Search result
Showing 1 - 50 of 4,696 items for (author: michael & g)

EMDB entry, No image
EMDB-43931: 
CryoEM structure of activated CRAF/MEK/14-3-3 complex with NST-628
Method: single particle / : Quade B, Cohen SE, Huang X

EMDB entry, No image
EMDB-43932: 
Activated CRAF/MEK heterotetramer from focused refinement of CRAF/MEK/14-3-3 complex
Method: single particle / : Quade B, Cohen SE, Huang X

PDB-9axa: 
CryoEM structure of activated CRAF/MEK/14-3-3 complex with NST-628
Method: single particle / : Quade B, Cohen SE, Huang X

PDB-9axc: 
Activated CRAF/MEK heterotetramer from focused refinement of CRAF/MEK/14-3-3 complex
Method: single particle / : Quade B, Cohen SE, Huang X

EMDB-19406: 
Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 12
Method: single particle / : Shilliday F, Lucas SCC, Richter M, Michaelides IN, Fusani L

EMDB-19407: 
Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 24
Method: single particle / : Shilliday F, Lucas SCC, Richter M, Michaelides IN, Fusani L

PDB-8rox: 
Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 12
Method: single particle / : Shilliday F, Lucas SCC, Richter M, Michaelides IN, Fusani L

PDB-8roy: 
Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 24
Method: single particle / : Shilliday F, Lucas SCC, Richter M, Michaelides IN, Fusani L

EMDB-43629: 
Cryo-EM structure of phage DEV ejection proteins gp72:gp73
Method: single particle / : Iglesias SM, Cingolani G

PDB-8vxq: 
Cryo-EM structure of phage DEV ejection proteins gp72:gp73
Method: single particle / : Iglesias SM, Cingolani G
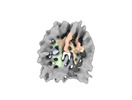
EMDB-19906: 
In situ nucleosome subtomogram average from Chlamydomonas reinhardtii
Method: subtomogram averaging / : Michael AK, Righetto RD, Obr M, van der Stappen P, Lamm L, Khavnekar S, Kotecha A, Engel BD

EMDB-41672: 
ELIC5 with Propylamine in spNW15 nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL

EMDB-41673: 
ELIC with Propylamine in spNW15 nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL

PDB-8twv: 
ELIC5 with Propylamine in spNW15 nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL

PDB-8twz: 
ELIC with Propylamine in spNW15 nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL

EMDB-40799: 
Cryo-EM consensus map of a double loaded human UBA7-UBE2L6-ISG15 thioester mimetic complex
Method: single particle / : Afsar M, Jia L, Ruben EA, Olsen SK

EMDB-42812: 
PNMA2 capsid, overall icosahedral map
Method: single particle / : Wilkinson ME, Madigan V, Zhang Y, Zhang F

EMDB-42815: 
PNMA2 capsid, focussed refinement of a pentamer (C5 symmetry)
Method: single particle / : Wilkinson ME, Madigan V, Zhang Y, Zhang F

PDB-8uyo: 
Structure of a recombinant human PNMA2 capsid
Method: single particle / : Wilkinson ME, Madigan V, Zhang Y, Zhang F

EMDB-17329: 
48S late-stage initiation complex with m6A mRNA
Method: single particle / : Guca E, Lima LHF, Boissier F, Hashem Y

EMDB-17330: 
48S late-stage initiation complex with non methylated mRNA
Method: single particle / : Guca E, Lima LHF, Boissier F, Hashem Y

PDB-8p03: 
48S late-stage initiation complex with m6A mRNA
Method: single particle / : Guca E, Lima LHF, Boissier F, Hashem Y

PDB-8p09: 
48S late-stage initiation complex with non methylated mRNA
Method: single particle / : Guca E, Lima LHF, Boissier F, Hashem Y

EMDB-19276: 
Cryo-EM of tetrameric collagenase-cleaved Xenopus ZP2-N2N3 (cleaved xZP2-N2N3) (C1)
Method: single particle / : Wiseman B, Nishio S, Jovine L

EMDB-19277: 
Cryo-EM of tetrameric collagenase-cleaved Xenopus ZP2-N2N3 (cleaved xZP2-N2N3) (C2)
Method: single particle / : Wiseman B, Nishio S, Jovine L
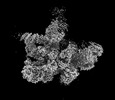
EMDB-18920: 
Plastid-encoded RNA polymerase (consensus map)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A
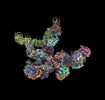
EMDB-18935: 
Plastid-encoded RNA polymerase
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18952: 
Plastid-encoded RNA polymerase transcription elongation complex (consensus map)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18964: 
Plastid-encoded RNA polymerase (Region 1)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18965: 
Plastid-encoded RNA polymerase (Region 2)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18974: 
Plastid-encoded RNA polymerase (Region 3)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18975: 
Plastid-encoded RNA polymerase (Region 4)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18976: 
Plastid-encoded RNA polymerase (Region 5)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18982: 
Plastid-encoded RNA polymerase (Region 6)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18983: 
Plastid-encoded RNA polymerase (Region 8)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18985: 
Plastid-encoded RNA polymerase (Region 9)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18986: 
Plastid-encoded RNA polymerase transcription elongation complex (Region 1)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18995: 
Plastid-encoded RNA polymerase transcription elongation complex (Region 2)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18996: 
Plastid-encoded RNA polymerase transcription elongation complex (Region 3)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18998: 
Plastid-encoded RNA polymerase (Region 7)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-19007: 
Plastid-encoded RNA polymerase transcription elongation complex (Region 4)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A
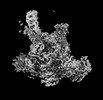
EMDB-19010: 
Plastid-encoded RNA polymerase transcription elongation complex (PAP2-mRNA)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

PDB-8r5o: 
Plastid-encoded RNA polymerase
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

PDB-8r6s: 
Plastid-encoded RNA polymerase (Integrated model)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

PDB-8rdj: 
Plastid-encoded RNA polymerase transcription elongation complex (Integrated model)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-41078: 
Acinetobacter baumannii 118362 family 2A cargo-loaded encapsulin shell
Method: single particle / : Andreas MP, Benisch R, Giessen TW

PDB-8t6r: 
Acinetobacter baumannii 118362 family 2A cargo-loaded encapsulin shell
Method: single particle / : Andreas MP, Benisch R, Giessen TW
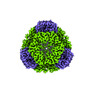
EMDB-42286: 
T33-ml28 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms
Method: single particle / : Castells-Graells R, Meador K, Sawaya MR, Yeates TO
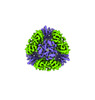
EMDB-42355: 
T33-ml30 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms
Method: single particle / : Castells-Graells R, Meador K, Sawaya MR, Yeates TO
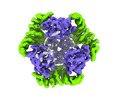
EMDB-42382: 
T33-ml35 Assembly Intermediate - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms
Method: single particle / : Castells-Graells R, Meador K, Sawaya MR, Yeates TO
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
