-Search query
-Search result
Showing all 39 items for (author: meyerson & jr)

EMDB-26719: 
FMC63 scFv in complex with soluble CD19
Method: single particle / : Meyerson J, He C

EMDB-26720: 
SJ25C1 Fab in complex with soluble CD19
Method: single particle / : Meyerson J, He C

EMDB-25414: 
Structure of human Kv1.3 with Fab-ShK fusion
Method: single particle / : Meyerson JR, Selvakumar P, Smider V, Huang R

EMDB-25416: 
Structure of human Kv1.3
Method: single particle / : Meyerson JR, Selvakumar P

EMDB-25417: 
Structure of human Kv1.3 with A0194009G09 nanobodies
Method: single particle / : Meyerson JR, Selvakumar P

EMDB-24444: 
Mouse GITR (mGITR) with DTA-1 Fab fragment
Method: single particle / : Meyerson JR, He C

EMDB-24789: 
Isoproterenol bound beta1 adrenergic receptor in complex with heterotrimeric Gi protein
Method: single particle / : Paknejad N, Alegre KO, Su M, Lou JS, Huang J, Jordan KD, Eng ET, Meyerson JR, Hite RK, Huang XY

EMDB-24790: 
Isoproterenol bound beta1 adrenergic receptor in complex with heterotrimeric Gi/s chimera protein
Method: single particle / : Paknejad N, Alegre KO, Su M, Lou JS, Huang J, Jordan KD, Eng ET, Meyerson JR, Hite RK, Huang XY

EMDB-23542: 
Structure of full-length GluK1 with L-Glu
Method: single particle / : Meyerson JR, Selvakumar P

EMDB-23494: 
Cryo-EM of the SLFN12-PDE3A complex: PDE3A body refinement
Method: single particle / : Fuller JR, Garvie CW, Lemke CT

EMDB-23495: 
Cryo-EM of the SLFN12-PDE3A complex: Consensus subset model
Method: single particle / : Fuller JR, Garvie CW, Lemke CT

EMDB-23496: 
Cryo-EM of the SLFN12-PDE3A complex: SLFN12 body refinement
Method: single particle / : Fuller JR, Garvie CW, Lemke CT

EMDB-23014: 
GluK2/K5 with 6-Cyano-7-nitroquinoxaline-2,3-dione (CNQX)
Method: single particle / : Khanra N, Brown PMGE, Perozzo AM, Bowie D, Meyerson JR

EMDB-23015: 
GluK2/K5 with L-Glu
Method: single particle / : Khanra N, Brown PMGE, Perozzo AM, Bowie D, Meyerson JR

EMDB-22357: 
Structural Basis of the Activation of Heterotrimeric Gs-protein by Isoproterenol-bound Beta1-Adrenergic Receptor
Method: single particle / : Su M, Zhu L, Zhang Y, Paknejad N, Dey R, Huang J, Lee MY, Williams D, Jordan KD, Eng ET, Ernst OP, Meyerson JR, Hite RK, Walz T, Liu W, Huang XY

EMDB-8289: 
GluK2EM with 2S,4R-4-methylglutamate
Method: single particle / : Meyerson JR, Chittori S, Merk A, Rao P, Han TH, Serpe M, Mayer ML, Subramaniam S

EMDB-8290: 
GluK2EM with LY466195
Method: single particle / : Meyerson JR, Chittori S

EMDB-2680: 
Density map of GluA2em in complex with ZK200775
Method: single particle / : Meyerson JR, Kumar J, Chittori S, Rao P, Pierson J, Bartesaghi A, Mayer ML, Subramaniam S

EMDB-2684: 
Density map of GluA2em in complex with LY451646 and glutamate
Method: single particle / : Meyerson JR, Kumar J, Chittori S, Rao P, Pierson J, Bartesaghi A, Mayer ML, Subramaniam S

EMDB-2685: 
Density map of GluK2 desensitized state in complex with 2S,4R-4-methylglutamate
Method: single particle / : Meyerson JR, Kumar J, Chittori S, Rao P, Pierson J, Bartesaghi A, Mayer ML, Subramaniam S

EMDB-2686: 
Density map of GluA2em desensitized state in complex with quisqualate (class 1)
Method: single particle / : Meyerson JR, Kumar J, Chittori S, Rao P, Pierson J, Bartesaghi A, Mayer ML, Subramaniam S

EMDB-2687: 
Density map of GluA2em desensitized state in complex with quisqualate (class 2)
Method: single particle / : Meyerson JR, Kumar J, Chittori S, Rao P, Pierson J, Bartesaghi A, Mayer ML, Subramaniam S

EMDB-2688: 
Density map of GluA2em desensitized state in complex with quisqualate (class 3)
Method: single particle / : Meyerson JR, Kumar J, Chittori S, Rao P, Pierson J, Bartesaghi A, Mayer ML, Subramaniam S

EMDB-2689: 
Density map of GluA2em in complex with quisqualate and LY451646
Method: single particle / : Meyerson JR, Kumar J, Chittori S, Rao P, Pierson J, Bartesaghi A, Mayer ML, Subramaniam S

EMDB-5682: 
Molecular structure of the native influenza hemagglutinin H1 on virions
Method: subtomogram averaging / : Harris AK, Meyerson JR, Matsuoka Y, Kuybeda O, Moran A, Bliss D, Das SR, Yewdell JW, Sapiro G, Subbarao K, Subramaniam S

EMDB-5683: 
Molecular structure of the native influenza hemagglutinin H1 on virions
Method: subtomogram averaging / : Harris AK, Meyerson JR, Matsuoka Y, Kuybeda O, Moran A, Bliss D, Das SR, Yewdell JW, Sapiro G, Subbarao K, Subramaniam S

EMDB-5684: 
Molecular structure of the native influenza hemagglutinin H1 on virions complexed with broadly neutralizing stem antibody C179
Method: subtomogram averaging / : Harris AK, Meyerson JR, Matsuoka Y, Kuybeda O, Moran A, Bliss D, Das SR, Yewdell JW, Sapiro G, Subbarao K, Subramaniam S

EMDB-5685: 
Molecular structure of the native influenza hemagglutinin H1 on virions complexed with broadly neutralizing stem antibody C179
Method: subtomogram averaging / : Harris AK, Meyerson JR, Matsuoka Y, Kuybeda O, Moran A, Bliss D, Das SR, Yewdell JW, Sapiro G, Subbarao K, Subramaniam S

EMDB-5686: 
Molecular structure of the native influenza hemagglutinin H3 on virions
Method: subtomogram averaging / : Harris AK, Meyerson JR, Matsuoka Y, Kuybeda O, Moran A, Bliss D, Das SR, Yewdell JW, Sapiro G, Subbarao K, Subramaniam S

EMDB-5687: 
Molecular structure of the native influenza hemagglutinin H3 on virions
Method: subtomogram averaging / : Harris AK, Meyerson JR, Matsuoka Y, Kuybeda O, Moran A, Bliss D, Das SR, Yewdell JW, Sapiro G, Subbarao K, Subramaniam S
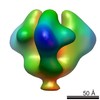
EMDB-5544: 
Molecular structure of the native HIV-1 Env trimer bound to VHH A12: Spike region
Method: subtomogram averaging / : Meyerson JR, Tran EEH, Kuybeda O, Chen W, Dimitrov DS, Gorlani A, Verrips T, Lifson JD, Subramaniam S

EMDB-5551: 
Molecular structure of the native HIV-1 Env trimer bound to VHH A12: Membrane region
Method: subtomogram averaging / : Meyerson JR, Tran EEH, Kuybeda O, Chen W, Dimitrov DS, Gorlani A, Verrips T, Lifson JD, Subramaniam S
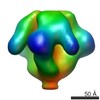
EMDB-5552: 
Molecular structure of the native HIV-1 Env trimer bound to m36: Spike region
Method: subtomogram averaging / : Meyerson JR, Tran EEH, Kuybeda O, Chen W, Dimitrov DS, Gorlani A, Verrips T, Lifson JD, Subramaniam S

EMDB-5553: 
Molecular structure of the native HIV-1 Env trimer bound to m36: Membrane region
Method: subtomogram averaging / : Meyerson JR, Tran EEH, Kuybeda O, Chen W, Dimitrov DS, Gorlani A, Verrips T, Lifson JD, Subramaniam S

EMDB-5554: 
Molecular structure of the native HIV-1 Env trimer bound to m36 and sCD4: Spike region
Method: subtomogram averaging / : Meyerson JR, Tran EEH, Kuybeda O, Chen W, Dimitrov DS, Gorlani A, Verrips T, Lifson JD, Subramaniam S

EMDB-5555: 
Molecular structure of the native HIV-1 Env trimer bound to m36 and sCD4: Membrane region
Method: subtomogram averaging / : Meyerson JR, Tran EEH, Kuybeda O, Chen W, Dimitrov DS, Gorlani A, Verrips T, Lifson JD, Subramaniam S

EMDB-5272: 
Molecular Structure of Unliganded Native SIVmneE11S gp120 trimer: Spike region
Method: subtomogram averaging / : White TA, Bartesaghi A, Borgnia M, Subramaniam S
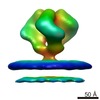
EMDB-5273: 
Molecular Structure of Unliganded Native SIVmac239 gp120 trimer: Spike region
Method: subtomogram averaging / : White TA, Bartesaghi A, Borgnia M, Subramaniam S

EMDB-5274: 
Molecular Structure of Unliganded Native CP-MAC gp120 trimer: Spike region
Method: subtomogram averaging / : White TA, Bartesaghi A, Borgnia M, Subramaniam S
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
