-Search query
-Search result
Showing all 50 items for (author: matsui & t)

EMDB-34530: 
Membrane protein A
Method: single particle / : Tajima S, Kim Y, Yamashita K, Fukuda M, Deisseroth K, Kato HE

EMDB-34531: 
Membrane protein B
Method: single particle / : Tajima S, Kim Y, Yamashita K, Fukuda M, Deisseroth K, Kato HE

EMDB-35713: 
Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 H225F mutant in lipid nanodisc
Method: single particle / : Tajima S, Kim Y, Nakamura S, Yamashita K, Fukuda M, Deisseroth K, Kato HE
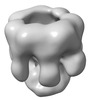
EMDB-32064: 
The oligomerized complex of hemolysin AF3 mutant protomers.
Method: single particle / : Ghanem N, Hashimoto T, Yokoyama T, Tanaka Y
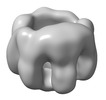
EMDB-32065: 
The oligomerized complex of AG2 hemolysin mutant protomers
Method: single particle / : Ghanem N, Hashimoto T, Yokoyama T, Tanaka Y

EMDB-32377: 
2.02 angstrom cryo-EM structure of the pump-like channelrhodopsin ChRmine
Method: single particle / : Kishi KE, Kim Y, Fukuda M, Yamashita K, Deisseroth K, Kato HE

EMDB-32378: 
2.12 angstrom cryo-EM map of the pump-like channelrhodopsin ChRmine with Fab antibody fragment
Method: single particle / : Kishi KE, Kim Y, Fukuda M, Yamashita K, Deisseroth K, Kato HE

PDB-7w9w: 
2.02 angstrom cryo-EM structure of the pump-like channelrhodopsin ChRmine
Method: single particle / : Kishi KE, Kim Y, Fukuda M, Yamashita K, Deisseroth K, Kato HE

EMDB-25046: 
Tomogram of mPCDH15/39G7-AuNP complex
Method: electron tomography / : Elferich J, Clark S, Ge J, Goehring A, Matsui A, Gouaux E

EMDB-25047: 
Tomogram of mouse stereocilia containing PCDH15 molecules labeled with 39G7-AuNPs
Method: electron tomography / : Elferich J, Clark S, Ge J, Goehring A, Matsui A, Gouaux E

EMDB-25048: 
Tomogram of mouse stereocilia containing PCDH15 molecules labeled with 39G7-AuNPs
Method: electron tomography / : Elferich J, Clark S, Ge J, Goehring A, Matsui A, Gouaux E

EMDB-25049: 
Tomogram of mouse stereocilia containing PCDH15 molecules labeled with 39G7-AuNPs
Method: electron tomography / : Elferich J, Clark S, Ge J, Goehring A, Matsui A, Gouaux E

EMDB-25050: 
Tomogram of mouse stereocilia containing PCDH15 molecules labeled with 39G7-AuNPs
Method: electron tomography / : Elferich J, Clark S, Ge J, Goehring A, Matsui A, Gouaux E

EMDB-25051: 
Tomogram of mouse stereocilia containing PCDH15 molecules labeled with 39G7-AuNPs
Method: electron tomography / : Elferich J, Clark S, Ge J, Goehring A, Matsui A, Gouaux E

EMDB-25052: 
Tomogram of mouse stereocilia containing PCDH15 molecules labeled with 39G7-AuNPs
Method: electron tomography / : Elferich J, Clark S, Ge J, Goehring A, Matsui A, Gouaux E

EMDB-25053: 
Tomogram of mouse stereocilia containing PCDH15 molecules labeled with 39G7-AuNPs
Method: electron tomography / : Elferich J, Clark S, Ge J, Goehring A, Matsui A, Gouaux E

EMDB-25054: 
Tomogram of mouse stereocilia containing PCDH15 molecules labeled with 39G7-AuNPs
Method: electron tomography / : Elferich J, Clark S, Ge J, Goehring A, Matsui A, Gouaux E

EMDB-25055: 
Tomogram of mouse stereocilia containing PCDH15 molecules labeled with 39G7-AuNPs
Method: electron tomography / : Elferich J, Clark S, Ge J, Goehring A, Matsui A, Gouaux E

EMDB-25056: 
Tomogram of mouse stereocilia containing PCDH15 molecules labeled with 39G7-AuNPs
Method: electron tomography / : Elferich J, Clark S, Ge J, Goehring A, Matsui A, Gouaux E

EMDB-25057: 
Tomogram of mouse stereocilia containing PCDH15 molecules labeled with 39G7-AuNPs
Method: electron tomography / : Elferich J, Clark S, Ge J, Goehring A, Matsui A, Gouaux E

EMDB-25058: 
Tomogram of mouse stereocilia containing PCDH15 molecules labeled with 39G7-AuNPs
Method: electron tomography / : Elferich J, Clark S, Ge J, Goehring A, Matsui A, Gouaux E

EMDB-25059: 
Tomogram of mouse stereocilia containing PCDH15 molecules labeled with 39G7-AuNPs
Method: electron tomography / : Elferich J, Clark S, Ge J, Goehring A, Matsui A, Gouaux E

EMDB-25060: 
Tomogram of mouse stereocilia containing PCDH15 molecules labeled with 39G7-AuNPs
Method: electron tomography / : Elferich J, Clark S, Ge J, Goehring A, Matsui A, Gouaux E

EMDB-25061: 
Tomogram of mouse stereocilia containing PCDH15 molecules labeled with 39G7-AuNPs
Method: electron tomography / : Elferich J, Clark S, Ge J, Goehring A, Matsui A, Gouaux E

EMDB-0999: 
Cryo-EM reconstruction of TLR7/Cpd-3 (SM-394830) complex in closed form
Method: single particle / : Zhang Z, Ohto U, Shimizu T

EMDB-1000: 
Cryo-EM reconstruction of TLR7/Cpd-6 (DSR-139293) complex in closed form
Method: single particle / : Zhang Z, Ohto U, Shimizu T

EMDB-30000: 
Cryo-EM reconstruction of TLR7/Cpd-6 (DSR-139293) complex in open form
Method: single particle / : Zhang Z, Ohto U, Shimizu T

EMDB-30001: 
Cryo-EM reconstruction of TLR7/Cpd-7 (DSR-139970) complex in open form
Method: single particle / : Zhang Z, Ohto U, Shimizu T

EMDB-30002: 
Cryo-EM structure of TLR7/Cpd-7 (DSR-139970) complex in open form
Method: single particle / : Zhang Z, Ohto U, Shimizu T

PDB-6lw1: 
Cryo-EM structure of TLR7/Cpd-7 (DSR-139970) complex in open form
Method: single particle / : Zhang Z, Ohto U, Shimizu T

EMDB-0809: 
Cryo-electron tomography of Candidatus Prometheoarchaeum syntrophicum strain MK-D1
Method: electron tomography / : Imachi H, Nobu MK, Nakahara N, Morono Y, Ogawara M, Takaki Y, Takano Y, Uematsu K, Ikuta T, Ito M, Matsui Y, Miyazaki M, Murata K, Saito Y, Sakai S, Song C, Tasumi E, Yamanaka Y, Yamaguchi T, Kamagata Y, Tamaki H, Takai K

EMDB-0852: 
Cryo-electron tomography of Candidatus Prometheoarchaeum syntrophicum strain MK-D1
Method: electron tomography / : Imachi H, Nobu MK, Nakahara N, Morono Y, Ogawara M, Takaki Y, Takano Y, Uematsu K, Ikuta T, Ito M, Matsui Y, Miyazaki M, Murata K, Saito Y, Sakai S, Song C, Tasumi E, Yamanaka Y, Yamaguchi T, Kamagata Y, Tamaki H, Takai K

EMDB-4750: 
CryoEM structure and molecular model of squid hemocyanin (Todarodes pacificus , TpH)
Method: single particle / : Tanaka Y, Kato S, Stabrin M, Raunser S, Matsui T, Gatsogiannis C

PDB-6r83: 
CryoEM structure and molecular model of squid hemocyanin (Todarodes pacificus , TpH)
Method: single particle / : Tanaka Y, Kato S, Stabrin M, Raunser S, Matsui T, Gatsogiannis C
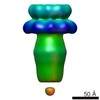
EMDB-6330: 
3D structure of the decameric assembly of the DNA-injection protein gp12 from bacteriophage Sf6
Method: single particle / : Zhao H, Speir JA, Matsui T, Lin Z, Liang L, Lynn AY, Varnado B, Weiss TM, Tang L

EMDB-5449: 
Dynamics in Cryo EM Reconstructions Visualized with Maximum-Likelihood Derived Variance Maps
Method: single particle / : Wang Q, Matsui T, Domitrovic T, Zheng Y, Doerschuk PC, Johnson JE

EMDB-5468: 
Dynamics in Cryo EM Reconstructions Visualized with Maximum-Likelihood Derived Variance Maps
Method: single particle / : Wang Q, Matsui T, Domitrovic T, Zheng Y, Doerschuk PC, Johnson JE

EMDB-5469: 
Dynamics in Cryo EM Reconstructions Visualized with Maximum-Likelihood Derived Variance Maps
Method: single particle / : Wang Q, Matsui T, Domitrovic T, Zheng Y, Doerschuk PC, Johnson JE

EMDB-5470: 
Dynamics in Cryo EM Reconstructions Visualized with Maximum-Likelihood Derived Variance Maps
Method: single particle / : Wang Q, Matsui T, Domitrovic T, Zheng Y, Doerschuk PC, Johnson JE

EMDB-5471: 
Dynamics in Cryo EM Reconstructions Visualized with Maximum-Likelihood Derived Variance Maps
Method: single particle / : Wang Q, Matsui T, Domitrovic T, Zheng Y, Doerschuk PC, Johnson JE

EMDB-5472: 
Dynamics in Cryo EM Reconstructions Visualized with Maximum-Likelihood Derived Variance Maps
Method: single particle / : Wang Q, Matsui T, Domitrovic T, Zheng Y, Doerschuk PC, Johnson JE

EMDB-5473: 
Dynamics in Cryo EM Reconstructions Visualized with Maximum-Likelihood Derived Variance Maps
Method: single particle / : Wang Q, Matsui T, Domitrovic T, Zheng Y, Doerschuk PC, Johnson JE

EMDB-5474: 
Dynamics in Cryo EM Reconstructions Visualized with Maximum-Likelihood Derived Variance Maps
Method: single particle / : Wang Q, Matsui T, Domitrovic T, Zheng Y, Doerschuk PC, Johnson JE

EMDB-5556: 
Negative stain electron microscopy structure of Nup192
Method: single particle / : Sampathkumar P, Kim SJ, Upla P, Rice W, Phillips J, Pieper U, Bonanno JB, Fernandez-Martinez J, Ketaren NE, Matsui T, Stokes DL, Sauder JM, Burley SK, Sali A, Rout MP, Almo SC

EMDB-5413: 
CryoEM map of the mature astrovirus (HAstV-1)
Method: single particle / : Dryden KA, Tihova M, Nowotny N, Matsui SM, Mendez E, Yeager M

EMDB-5414: 
CryoEM map of the immature human astrovirus HAstV-8
Method: single particle / : Dryden KA, Tihova M, Nowotny N, Matsui SM, Mendez E, Yeager M

EMDB-5425: 
Cryo-EM reconstruction of fully-mature capsid of Nudaurelia capensis omega virus (NwV)
Method: single particle / : Matsui T, Lander GC, Khayat R, Johnson JE

EMDB-5426: 
Cryo-EM reconstruction of Nudaurelia capensis omega virus (NwV) capsid at 3min time point following maturation initiation
Method: single particle / : Matsui T, Lander GC, Khayat R, Johnson JE

EMDB-5427: 
Cryo-EM reconstruction of Nudaurelia capensis omega virus (NwV) capsid at 30min time point following maturation initiation
Method: single particle / : Matsui T, Lander GC, Khayat R, Johnson JE

EMDB-5428: 
Cryo-EM reconstruction of Nudaurelia capensis omega virus (NwV) capsid at 4hr time point following maturation initiation
Method: single particle / : Matsui T, Lander GC, Khayat R, Johnson JE
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
