-Search query
-Search result
Showing 1 - 50 of 267 items for (author: martin & tg)

EMDB-27973: 
Erwinia amylovora 70S Ribosome
Method: subtomogram averaging / : Laughlin TG, Villa E

EMDB-27993: 
Erwinia amlyavora 50S ribosome
Method: subtomogram averaging / : Prichard A, Lee J, Laughlin TG, Lee A, Thomas K, Sy A, Spencer T, Asavavimol A, Cafferata A, Cameron M, Chiu N, Davydov D, Desai I, Diaz G, Guereca M, Hearst K, Huang L, Jacobs E, Johnson A, Kahn S, Koch R, Martinez A, McDonald-Uyeno A, Norquist M, Pau T, Prasad G, Saam K, Sandu M, Sarabia A, Schumaker S, Sonin A, Zhao A, Corbett A, Pogliano K, Meyer J, Grose J, Villa E, Dutton R, Pogliano J

EMDB-28003: 
Erwinia phage vB_EamM_RAY (RAY) Capsid Vertex
Method: subtomogram averaging / : Laughlin TG, Villa E

EMDB-28004: 
Erwinia phage vB_EamM_RAY (RAY) Capsid Collar
Method: subtomogram averaging / : Laughlin TG, Villa E

EMDB-28005: 
Erwinia phage vB_EamM_RAY (RAY) Tail Sheath
Method: subtomogram averaging / : Laughlin TG, Villa E

EMDB-28006: 
Erwinia phage vB_EamM_RAY (RAY) Baseplate
Method: subtomogram averaging / : Laughlin TG, Villa E

EMDB-28007: 
Erwinia phage vB_EamM_RAY (RAY) Chimallin
Method: subtomogram averaging / : Laughlin TG, Villa E

EMDB-28008: 
Erwinia phage vB_EamM_RAY (RAY) Putative PhuZ Filament
Method: subtomogram averaging / : Laughlin TG, Villa E

EMDB-28009: 
Pseudomonas chlororaphis phage 201phi2-1 PhuZ Filament
Method: subtomogram averaging / : Laughlin TG, Villa E

EMDB-28010: 
Pseudomonas phage phiKZ PhuZ Filament
Method: subtomogram averaging / : Laughlin TG, Villa E
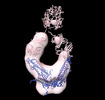
EMDB-26217: 
Negative stain EM map of COVA1-07 mAb bound to the S2 domain of SARS-CoV-2 S
Method: single particle / : Han J, Ward AB
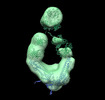
EMDB-26218: 
Negative stain EM map of COVA2-14 mAb bound to the S2 domain of SARS-CoV-2 S
Method: single particle / : Han J, Ward AB

EMDB-26219: 
Negative stain EM map of COVA2-18 mAb bound to the S2 domain of SARS-CoV-2 S
Method: single particle / : Han J, Ward AB
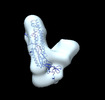
EMDB-26220: 
Negative stain EM map of the S2 domain of SARS-CoV-2 S
Method: single particle / : Han J, Ward AB

EMDB-13326: 
T. maritima CorA in DDM micelles without Mg2+ bound in D2O
Method: single particle / : Larsen AH, Johansen NT, Arleth L

EMDB-13327: 
T. maritima CorA in DDM micelles with Mg2+ bound in D2O
Method: single particle / : Larsen AH, Johansen NT, Arleth L

EMDB-13290: 
a1b3 GABA-A receptor + GABA
Method: single particle / : Miller PS, Kasaragod VB, Hardwick SW, Chirgadze DY

EMDB-13314: 
a1b3 GABA-A receptor + GABA
Method: single particle / : Miller PS, Kasaragod VB, Hardwick SW, Chirgadze DY

EMDB-13315: 
GABA-A receptor bound by a-Cobratoxin
Method: single particle / : Kasaragod VB, Hardwick SW, Chirgadze DY, Miller PS

EMDB-25105: 
Structure of SARS-CoV S protein in complex with Receptor Binding Domain antibody DH1047
Method: single particle / : Gobeil S, Acharya P

PDB-7sg4: 
Structure of SARS-CoV S protein in complex with Receptor Binding Domain antibody DH1047
Method: single particle / : Gobeil S, Acharya P

EMDB-12378: 
Mycobacterium smegmatis ATP synthase F1 state 1a
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12379: 
Mycobacterium smegmatis ATP synthase Fo state 1a
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12380: 
Mycobacterium smegmatis ATP synthase Peripheral Stalk state 1a
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12381: 
Mycobacterium smegmatis ATP synthase b-delta state 1a
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12383: 
Mycobacterium smegmatis ATP synthase F1 state 1b
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12384: 
Mycobacterium smegmatis ATP synthase Fo state 1b
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12385: 
Mycobacterium smegmatis ATP synthase Peripheral Stalk state 1b
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12386: 
Mycobacterium smegmatis ATP synthase b-delta state 1b
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12388: 
Mycobacterium smegmatis ATP synthase F1 state 1c
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12389: 
Mycobacterium smegmatis ATP synthase Fo state 1c
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12390: 
Mycobacterium smegmatis ATP synthase Peripheral Stalk state 1c
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12391: 
Mycobacterium smegmatis ATP synthase b-delta state 1c
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12393: 
Mycobacterium smegmatis ATP synthase F1 state 1d
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12394: 
Mycobacterium smegmatis ATP synthase Fo state 1d
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12395: 
Mycobacterium smegmatis ATP synthase Peripheral Stalk state 1d
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12396: 
Mycobacterium smegmatis ATP synthase b-delta state 1d
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12398: 
Mycobacterium smegmatis ATP synthase F1 state 1e
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12399: 
Mycobacterium smegmatis ATP synthase Fo state 1e
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12400: 
Mycobacterium smegmatis ATP synthase Peripheral Stalk state 1e
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12401: 
Mycobacterium smegmatis ATP synthase b-delta state 1e
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12403: 
Mycobacterium smegmatis ATP synthase F1 state 2
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12405: 
Mycobacterium smegmatis ATP synthase Peripheral Stalk state 2
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12408: 
Mycobacterium smegmatis ATP synthase F1 state 3a
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12409: 
Mycobacterium smegmatis ATP synthase Fo state 3a
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12410: 
Mycobacterium smegmatis ATP synthase Peripheral Stalk state 3a
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12411: 
Mycobacterium smegmatis ATP synthase b-delta state 3a
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12413: 
Mycobacterium smegmatis ATP synthase F1 state 3b
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12414: 
Mycobacterium smegmatis ATP synthase Fo state 3b
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE

EMDB-12415: 
Mycobacterium smegmatis ATP synthase Peripheral Stalk state 3b
Method: single particle / : Petri J, Montgomery MG, Spikes TE, Walker JE
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
