-Search query
-Search result
Showing all 48 items for (author: mark & r & h & hurst)

EMDB-18953: 
Cryo-electron tomogram of Yersinia entomophaga chi2-sfGFP cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-18954: 
Cryo-electron tomogram of Yersinia entomophaga chi2-sfGFP cells
Method: electron tomography / : Feldmueller M, Pilhofer M
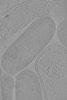
EMDB-18955: 
Cryo-electron tomogram of Yersinia entomophaga chi2-sfGFP cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-18957: 
Cryo-electron tomogram of Yersinia entomophaga MH96 cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-18958: 
Cryo-electron tomogram of Yersinia entomophaga MH96 cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-18960: 
Cryo-electron tomogram of Yersinia entomophaga delta LC cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-18961: 
Cryo-electron tomogram of mechanically cryo-milled Yersinia entomophaga delta LC cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-18962: 
Cryo-electron tomogram of a lysate preparation of Yersinia entomophaga delta LC cells
Method: electron tomography / : Feldmueller M, Pilhofer M
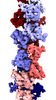
EMDB-18970: 
Subtomogram average of M66 filaments in Yersinia entomophaga cells
Method: subtomogram averaging / : Feldmueller M, Afanasyev P, Pilhofer M
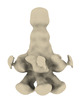
EMDB-18971: 
Subtomogram average of YenTc-Chi2-sfGFP from Yersinia entomophaga chi2-sfGFP
Method: subtomogram averaging / : Feldmueller M, Pilhofer M
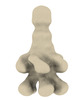
EMDB-18972: 
Subtomogram average of YenTc from Yersinia entomophaga MH96
Method: subtomogram averaging / : Feldmueller M, Pilhofer M

EMDB-19370: 
Cryo-electron tomogram of Yersinia entomophaga delta YenTc cells
Method: electron tomography / : Feldmueller M, Pilhofer M
EMDB-19371: 
Cryo-electron tomogram of Yersinia entomophaga delta YenTc cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19372: 
Cryo-electron tomogram of Yersinia entomophaga delta YenTc cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19373: 
Cryo-electron tomogram of Yersinia entomophaga delta YenTc cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19374: 
Cryo-electron tomogram of Yersinia entomophaga chi2-sfGFP cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19375: 
Cryo-electron tomogram of Yersinia entomophaga chi2-sfGFP cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19376: 
Cryo-electron tomogram of Yersinia entomophaga delta LC delta YenTc cells
Method: electron tomography / : Feldmueller M, Pilhofer M
EMDB-19377: 
Cryo-electron tomogram of Yersinia entomophaga MH96 cells grown at 37 degrees
Method: electron tomography / : Feldmueller M, Pilhofer M
EMDB-19378: 
Cryo-electron tomogram of Yersinia entomophaga delta LC cells grown at 37 degrees
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19379: 
Cryo-electron tomogram of Yersinia entomophaga delta LC delta M66 cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19380: 
Cryo-electron tomogram of Yersinia entomophaga delta LC delta M66 cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19381: 
Cryo-electron tomogram of a lysate preparation of Yersinia entomophaga delta LC delta M66 cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-11997: 
Structure of a nanoparticle for a COVID-19 vaccine candidate
Method: single particle / : Duyvesteyn HME, Stuart DI

PDB-7b3y: 
Structure of a nanoparticle for a COVID-19 vaccine candidate
Method: single particle / : Duyvesteyn HME, Stuart DI

EMDB-20053: 
Cryo-EM structure of YenTcA in its prepore state
Method: single particle / : Piper SJ, Brillault L

EMDB-20054: 
Cryo-EM structure of YenTcA in the pore state determined in liposomes
Method: single particle / : Landsberg MJ, Piper SJ

PDB-6ogd: 
Cryo-EM structure of YenTcA in its prepore state
Method: single particle / : Piper SJ, Brillault L, Box JK, Landsberg MJ

EMDB-4800: 
Cryo-EM structure of the anti-feeding prophage (AFP) baseplate in extended state, 3-fold symmetrised
Method: single particle / : Desfosses A

EMDB-4802: 
Cryo-EM structure of the anti-feeding prophage (AFP) helical sheath-tube complex in extended state
Method: helical / : Desfosses A

EMDB-4859: 
Cryo-EM structure of the anti-feeding prophage (AFP) sheath-tube in contracted state, C6 symmetrized
Method: single particle / : Desfosses A

EMDB-4871: 
Cryo-EM structure of the anti-feeding prophage (AFP) needle from signal-subtracted particles
Method: single particle / : Desfosses A

EMDB-4876: 
Cryo-EM structure of the anti-feeding prophage (AFP) baseplate in contracted state
Method: single particle / : Desfosses A

PDB-6rbk: 
Cryo-EM structure of the anti-feeding prophage (AFP) baseplate in extended state, 3-fold symmetrised
Method: single particle / : Desfosses A

PDB-6rbn: 
Cryo-EM structure of the anti-feeding prophage (AFP) helical sheath-tube complex in extended state
Method: helical / : Desfosses A

PDB-6rgl: 
Cryo-EM structure of the anti-feeding prophage (AFP) baseplate in contracted state
Method: single particle / : Desfosses A

EMDB-4782: 
Cryo-EM structure of the anti-feeding prophage (AFP) baseplate, 6-fold symmetrised
Method: single particle / : Desfosses A

EMDB-4783: 
Composite map of an entire contractile injection device : the anti-feeding prophage (AFP)
Method: single particle / : Desfosses A

EMDB-4784: 
Cryo-EM structure of the anti-feeding prophage cap (AFP tube terminating cap)
Method: single particle / : Desfosses A

EMDB-4801: 
Cryo-EM structure of the anti-feeding prophage cap (AFP tube terminating cap), ending with Afp3
Method: single particle / : Desfosses A

EMDB-4803: 
Cryo-EM structure of the anti-feeding prophage (AFP) helical sheath in contracted state
Method: helical / : Desfosses A

PDB-6rao: 
Cryo-EM structure of the anti-feeding prophage (AFP) baseplate, 6-fold symmetrised
Method: single particle / : Desfosses A

PDB-6rap: 
Cryo-EM structure of the anti-feeding prophage cap (AFP tube terminating cap)
Method: single particle / : Desfosses A

PDB-6rc8: 
Cryo-EM structure of the anti-feeding prophage (AFP) helical sheath in contracted state
Method: helical / : Desfosses A

EMDB-2419: 
3-dimensional structure of the toxin-delivery particle antifeeding prophage of Serratia entomophila
Method: single particle / : Heymann JB, Bartho JD, Rybakova D, Venugopal HP, Winkler DC, Sen A, Hurst MRH, Mitra AK

EMDB-2423: 
3-dimensional structure of the toxin-delivery particle antifeeding prophage of Serratia entomophila
Method: single particle / : Heymann JB, Bartho JD, Rybakova D, Venugopal HP, Winkler DC, Sen A, Hurst MRH, Mitra AK

EMDB-1978: 
3D structure of the Yersinia entomophaga toxin complex and implications for insecticidal activity
Method: single particle / : Landsberg MJ, Jones SA, Rothnagel R, Busby JN, Marshall SDG, Simpson RM, Lott JS, Hankamer B, Hurst MRH

PDB-4a5q: 
Crystal structure of the chitinase Chi1 fitted into the 3D structure of the Yersinia entomophaga toxin complex
Method: single particle / : Busby JN, Landsberg MJ, Simpson RM, Jones SA, Hankamer B, Hurst MRH, Lott JS
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
