-Search query
-Search result
Showing 1 - 50 of 2,471 items for (author: mark & a)
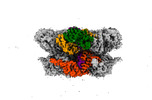
EMDB-42603: 
Human p97/VCP structure with a triazole inhibitor (NSC799462/hexamer)
Method: single particle / : Nandi P, DeVore K, Chiu PL
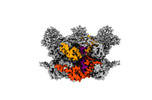
EMDB-42625: 
Human p97/VCP R155H mutant structure with a triazole inhibitor (NSC804515)
Method: single particle / : Nandi P, DeVore K, Chiu PL
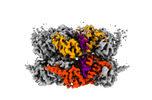
EMDB-42626: 
Human p97/VCP R155H mutant structure with a triazole inhibitor (NSC819701/up)
Method: single particle / : Nandi P, DeVore K, Chiu PL
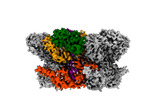
EMDB-42627: 
Human p97/VCP R155H mutant structure with a triazole inhibitor (NSC819701/down)
Method: single particle / : Nandi P, DeVore K, Chiu PL
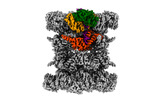
EMDB-44748: 
Human p97/VCP structure with a triazole inhibitor (NSC799462/dodecamer)
Method: single particle / : Nandi P, DeVore K, Chiu PL
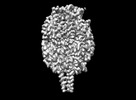
EMDB-43516: 
Cryo-EM structure of HMPV (MPV-2cREKR)
Method: single particle / : Yu X, Langedijk JPM

EMDB-43517: 
Cryo-EM structure of HMPV (MPV-2cREKR)
Method: single particle / : Yu X, Langedijk JPM

PDB-8vt2: 
cryo-EM structure of HMPV (MPV-2c)
Method: single particle / : Yu X, Langedijk JPM

PDB-8vt3: 
cryo-EM structure of HMPV (MPV-2cREKR)
Method: single particle / : Yu X, Langedijk JPM

EMDB-19822: 
Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture +PIP2/+bromosterol (DOPC, DOPE, DOPS, bromo-ergosterol, PI(4,5)P2 35:20:20:15:10)
Method: helical / : Kefauver JM, Zou L, Desfosses A, Loewith RJ

EMDB-42400: 
RORC mRNA 3'UTR riboswitch A97G/G98A mutant class C
Method: single particle / : Asarnow D, Khoroshkin M, Goodarzi H, Cheng Y

EMDB-42401: 
RORC mRNA 3'UTR riboswitch 77-GA mutant class A
Method: single particle / : Asarnow D, Khoroshkin M, Goodarzi H, Cheng Y

EMDB-42403: 
RORC mRNA 3'UTR riboswitch 117-AC mutant class C
Method: single particle / : Asarnow D, Khoroshkin M, Goodarzi H, Cheng Y

EMDB-42404: 
RORC mRNA 3'UTR riboswitch 117-AC mutant class B
Method: single particle / : Asarnow D, Khoroshkin M, Goodarzi H, Cheng Y

EMDB-40218: 
CryoEM structure of TnsC(1-503) bound to TnsD(1-318) from E.coli Tn7
Method: single particle / : Shen Y, Guarne A

EMDB-40221: 
CryoEM structure of the TnsC(1-503)-TnsD(1-318)-DNA complex in a 7:2:1 stoichiometry from E. coli Tn7
Method: single particle / : Shen Y, Guarne A

EMDB-40222: 
CryoEM structure of the TnsC(1-503)-TnsD(1-318)-DNA complex in a 6:2:1 stoichiometry from E. coli Tn7
Method: single particle / : Shen Y, Guarne A

EMDB-43138: 
CryoEM structure of the TnsC(1-503)-TnsD(1-318)-DNA complex in a 7:2:1 stoichiometry from E. coli Tn7 bound to ATPgS and ADP
Method: single particle / : Shen Y, Guarne A

EMDB-43140: 
CyoEM structure of the TnsC(1-503)-TnsD(1-318)-DNA complex in a 6:2:1 stoichiometry from E. coli Tn7 bound to ATPgS and ADP
Method: single particle / : Shen Y, Guarne A

PDB-8glu: 
CryoEM structure of TnsC(1-503) bound to TnsD(1-318) from E.coli Tn7
Method: single particle / : Shen Y, Guarne A

PDB-8glw: 
CryoEM structure of the TnsC(1-503)-TnsD(1-318)-DNA complex in a 7:2:1 stoichiometry from E. coli Tn7
Method: single particle / : Shen Y, Guarne A

PDB-8glx: 
CryoEM structure of the TnsC(1-503)-TnsD(1-318)-DNA complex in a 6:2:1 stoichiometry from E. coli Tn7
Method: single particle / : Shen Y, Guarne A

PDB-8vcj: 
CryoEM structure of the TnsC(1-503)-TnsD(1-318)-DNA complex in a 7:2:1 stoichiometry from E. coli Tn7 bound to ATPgS and ADP
Method: single particle / : Shen Y, Guarne A

PDB-8vct: 
CyoEM structure of the TnsC(1-503)-TnsD(1-318)-DNA complex in a 6:2:1 stoichiometry from E. coli Tn7 bound to ATPgS and ADP
Method: single particle / : Shen Y, Guarne A

EMDB-18307: 
Native eisosome lattice bound to plasma membrane microdomain
Method: single particle / : Kefauver JM, Zou L, Loewith RJ, Defosses A

EMDB-18308: 
Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture -PIP2/+sterol (DOPC, DOPE, DOPS, cholesterol 30:20:20:30)
Method: helical / : Kefauver JM, Zou L, Desfosses A, Loewith RJ

EMDB-18309: 
Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture +PIP2/-sterol (DOPC, DOPE, DOPS, PI(4,5)P2 50:20:20:10)
Method: helical / : Kefauver JM, Zou L, Desfosses A, Loewith RJ

EMDB-18310: 
Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture +PIP2/+sterol (DOPC, DOPE, DOPS, cholesterol, PI(4,5)P2 35:20:20:15:10)
Method: helical / : Kefauver JM, Zou L, Desfosses A, Loewith RJ

EMDB-18311: 
Compact state - Native eisosome lattice bound to plasma membrane microdomain
Method: single particle / : Kefauver JM, Zou L, Desfosses A, Loewith RJ

EMDB-18312: 
Stretched state - Native eisosome lattice bound to plasma membrane microdomain
Method: single particle / : Kefauver JM, Zou L, Desfosses A, Loewith RJ

PDB-8qb7: 
Pil1 in native eisosome lattice bound to plasma membrane microdomain
Method: single particle / : Kefauver JM, Zou L, Loewith RJ, Defosses A

PDB-8qb8: 
Lsp1 in native eisosome lattice bound to plasma membrane microdomain
Method: single particle / : Kefauver JM, Zou L, Loewith RJ, Defosses A

PDB-8qb9: 
Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture -PIP2/+sterol (DOPC, DOPE, DOPS, cholesterol 30:20:20:30)
Method: helical / : Kefauver JM, Zou L, Desfosses A, Loewith RJ

PDB-8qbb: 
Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture +PIP2/-sterol (DOPC, DOPE, DOPS, PI(4,5)P2 50:20:20:10)
Method: helical / : Kefauver JM, Zou L, Desfosses A, Loewith RJ

PDB-8qbd: 
Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture +PIP2/+sterol (DOPC, DOPE, DOPS, cholesterol, PI(4,5)P2 35:20:20:15:10)
Method: helical / : Kefauver JM, Zou L, Desfosses A, Loewith RJ

PDB-8qbe: 
Compact state - Pil1 in native eisosome lattice bound to plasma membrane microdomain
Method: single particle / : Kefauver JM, Zou L, Desfosses A, Loewith RJ

PDB-8qbf: 
Compact state - Pil1 dimer with lipid headgroups fitted in native eisosome lattice bound to plasma membrane microdomain
Method: single particle / : Kefauver JM, Zou L, Desfosses A, Loewith RJ

PDB-8qbg: 
Stretched state - Pil1 in native eisosome lattice bound to plasma membrane microdomain
Method: single particle / : Kefauver JM, Zou L, Desfosses A, Loewith RJ

EMDB-18180: 
cryoEM structure of SARS-CoV2 Spike trimer in complex with Fab23
Method: single particle / : Hallberg M, Das H

PDB-8q5y: 
cryoEM structure of SARS-CoV2 Spike trimer in complex with Fab23
Method: single particle / : Hallberg M, Das H

EMDB-18402: 
cryo-EM structure of apo-TcdB
Method: single particle / : Kinsolving J, Bous J

EMDB-18403: 
cryo-EM map of apo Clostridioides difficile toxin B
Method: single particle / : Kinsolving J, Bous J

EMDB-18409: 
cryo-EM structure of TcdB-FZD7
Method: single particle / : Kinsolving J, Bous J

EMDB-18410: 
cryo-EM structure of TcdB-FZD7
Method: single particle / : Kinsolving J, Bous J

EMDB-18411: 
cryo-EM structure of TcdB-FZD7
Method: single particle / : Kinsolving J, Bous J

EMDB-50814: 
Real space helical reconstruction of cofilin actin in the microtubule lumen of human platelets
Method: helical / : Tsuji C, Bradshaw M, Paul DM, Dodding MP

EMDB-50845: 
Real space helical reconstruction of cofilin actin in the microtubule lumen of HAP1 cells
Method: helical / : Tsuji C, Bradshaw M, Paul DM, Dodding MP

EMDB-43296: 
Constituent EM map: Focused refinement S100A1 of mouse RyR1 in complex with S100A1 (EGTA-only dataset)
Method: single particle / : Weninger G, Marks AR

EMDB-18438: 
mt-SSU assembly intermediate in GTPBP8 knock-out cells, state 1
Method: single particle / : Valentin Gese G, Cipullo M, Rorbach J, Hallberg BM

EMDB-18439: 
mt-SSU assembly intermediate in GTPBP8 knock-out cells, state 2
Method: single particle / : Valentin Gese G, Cipullo M, Rorbach J, Hallberg BM
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
