-Search query
-Search result
Showing all 46 items for (author: macdonald & a)

EMDB-18268: 
P301S Tau Filaments from the Brains of PS19 Transgenic Mouse Line
Method: helical / : Schweighauser M, Murzin AG, Macdonald J, Lavenir I, Crowther RA, Scheres SHW, Goedert M

EMDB-18269: 
P301S Tau Filaments from the Brains of Tg2541 Transgenic Mouse Line
Method: helical / : Schweighauser M, Murzin AG, Macdonald J, Lavenir I, Crowther RA, Scheres SHW, Goedert M

PDB-8q92: 
P301S Tau Filaments from the Brains of PS19 Transgenic Mouse Line
Method: helical / : Schweighauser M, Murzin AG, Macdonald J, Lavenir I, Crowther RA, Scheres SHW, Goedert M

PDB-8q96: 
P301S Tau Filaments from the Brains of Tg2541 Transgenic Mouse Line
Method: helical / : Schweighauser M, Murzin AG, Macdonald J, Lavenir I, Crowther RA, Scheres SHW, Goedert M

EMDB-16022: 
Amyloid-beta 42 filaments extracted from the human brain with Arctic mutation (E22G) of Alzheimer's disease | ABeta42
Method: helical / : Yang Y, Zhang WJ, Murzin AG, Schweighauser M, Huang M, Lovestam SKA, Peak-Chew SY, Macdonald J, Lavenir I, Ghetti B, Graff C, Kumar A, Nordberg A, Goedert M, Scheres SHW

EMDB-16023: 
Amyloid-beta tetrameric filaments with the Arctic mutation (E22G) from Alzheimer's disease brains | ABeta40
Method: helical / : Yang Y, Zhang WJ, Murzin AG, Schweighauser M, Huang M, Lovestam SKA, Peak-Chew SY, Macdonald J, Lavenir I, Ghetti B, Graff C, Kumar A, Nordber A, Goedert M, Scheres SHW

EMDB-16027: 
Murine amyloid-beta filaments with the Arctic mutation (E22G) from APP(NL-G-F) mouse brains | ABeta
Method: helical / : Yang Y, Zhang WJ, Murzin AG, Schweighauser M, Huang M, Lovestam SKA, Peak-Chew SY, Macdonald J, Lavenir I, Ghetti B, Graff C, Kumar A, Nordber A, Goedert M, Scheres SHW

PDB-8bfz: 
Amyloid-beta 42 filaments extracted from the human brain with Arctic mutation (E22G) of Alzheimer's disease | ABeta42
Method: helical / : Yang Y, Zhang WJ, Murzin AG, Schweighauser M, Huang M, Lovestam SKA, Peak-Chew SY, Macdonald J, Lavenir I, Ghetti B, Graff C, Kumar A, Nordberg A, Goedert M, Scheres SHW

PDB-8bg0: 
Amyloid-beta tetrameric filaments with the Arctic mutation (E22G) from Alzheimer's disease brains | ABeta40
Method: helical / : Yang Y, Zhang WJ, Murzin AG, Schweighauser M, Huang M, Lovestam SKA, Peak-Chew SY, Macdonald J, Lavenir I, Ghetti B, Graff C, Kumar A, Nordber A, Goedert M, Scheres SHW

PDB-8bg9: 
Murine amyloid-beta filaments with the Arctic mutation (E22G) from APP(NL-G-F) mouse brains | ABeta
Method: helical / : Yang Y, Zhang WJ, Murzin AG, Schweighauser M, Huang M, Lovestam SKA, Peak-Chew SY, Macdonald J, Lavenir I, Ghetti B, Graff C, Kumar A, Nordber A, Goedert M, Scheres SHW

EMDB-16434: 
Type Ib beta-amyloid 42 Filaments from Human Brain
Method: helical / : Yang Y, Arseni D, Zhang W, Huang M, Lovestam SKA, Schweighauser M, Kotecha A, Murzin AG, Peak-Chew SY, Macdonald J, Lavenir I, Garringer HJ, Gelpi E, Newell KL, Kovacs GG, Vidal R, Ghetti B, Falcon B, Scheres HW, Goedert M

EMDB-25476: 
G32A4 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction)
Method: single particle / : Windsor IW, Tong P, Wesemann DR, Harrison SC

EMDB-25477: 
C98C7 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction)
Method: single particle / : Windsor IW, Tong P, Wesemann DR, Harrison SC

EMDB-25478: 
G32Q4 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction)
Method: single particle / : Windsor IW, Tong P, Wesemann DR, Harrison SC

PDB-7swn: 
G32A4 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction)
Method: single particle / : Windsor IW, Tong P, Wesemann DR, Harrison SC

PDB-7swo: 
C98C7 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction)
Method: single particle / : Windsor IW, Tong P, Wesemann DR, Harrison SC

PDB-7swp: 
G32Q4 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction)
Method: single particle / : Windsor IW, Tong P, Wesemann DR, Harrison SC

EMDB-13800: 
Type I beta-amyloid 42 Filaments from Human Brain
Method: helical / : Yang Y, Arseni D, Zhang W, Huang M, Lovestam SKA, Schweighauser M, Kotecha A, Murzin AG, Peak-Chew SY, Macdonald J, Lavenir I, Garringer HJ, Gelpi E, Newell KL, Kovacs GG, Vidal R, Ghetti B, Falcon B, Scheres SHW, Goedert M

EMDB-13809: 
Type II beta-amyloid 42 Filaments from Human Brain
Method: helical / : Yang Y, Arseni D, Zhang W, Huang M, Lovestam SKA, Schweighauser M, Kotecha A, Murzin AG, Peak-Chew SY, Macdonald J, Lavenir I, Garringer HJ, Gelpi E, Newell KL, Kovacs GG, Vidal R, Ghetti B, Falcon B, Scheres SHW, Goedert M
PDB-7q4b: 
Type I beta-amyloid 42 Filaments from Human Brain
Method: helical / : Yang Y, Arseni D, Zhang W, Huang M, Lovestam SKA, Schweighauser M, Kotecha A, Murzin AG, Peak-Chew SY, Macdonald J, Lavenir I, Garringer HJ, Gelpi E, Newell KL, Kovacs GG, Vidal R, Ghetti B, Falcon B, Scheres SHW, Goedert M

PDB-7q4m: 
Type II beta-amyloid 42 Filaments from Human Brain
Method: helical / : Yang Y, Arseni D, Zhang W, Huang M, Lovestam SKA, Schweighauser M, Kotecha A, Murzin AG, Peak-Chew SY, Macdonald J, Lavenir I, Garringer HJ, Gelpi E, Newell KL, Kovacs GG, Vidal R, Ghetti B, Falcon B, Scheres SHW, Goedert M

EMDB-23280: 
Cryo-EM structure of the human adenosine A1 receptor-Gi2-protein complex bound to its endogenous agonist and an allosteric ligand
Method: single particle / : Draper-Joyce CJ, Danev R, Thal DM, Christopoulos A, Glukhova A

EMDB-23281: 
Cryo-EM structure of the human adenosine A1 receptor-Gi2-protein complex bound to its endogenous agonist
Method: single particle / : Draper-Joyce CJ, Danev R, Thal DM, Christopoulos A, Glukhova A

PDB-7ld3: 
Cryo-EM structure of the human adenosine A1 receptor-Gi2-protein complex bound to its endogenous agonist and an allosteric ligand
Method: single particle / : Draper-Joyce CJ, Danev R, Thal DM, Christopoulos A, Glukhova A

PDB-7ld4: 
Cryo-EM structure of the human adenosine A1 receptor-Gi2-protein complex bound to its endogenous agonist
Method: single particle / : Draper-Joyce CJ, Danev R, Thal DM, Christopoulos A, Glukhova A

EMDB-24194: 
SARS-CoV-2 Spike (2P) in complex with C12C11 Fab
Method: single particle / : Windsor IW, Bajic G, Tong P, Gautam AK, Wesemann DR, Harrison SC

EMDB-24192: 
SARS-CoV-2 Spike (2P) in complex with C12C9 Fab (NTD local reconstruction)
Method: single particle / : Windsor IW, Jenni S, Bajic G, Tong P, Gautam AK, Wesemann DR, Harrison SC

EMDB-24193: 
SARS-CoV-2 Spike (2P) in complex with G32R7 Fab (RBD and NTD local reconstruction)
Method: single particle / : Windsor IW, Jenni S, Tong P, Gautam AK, Wesemann DR, Harrison SC

EMDB-24196: 
SARS-CoV-2 spike (6P) in complex with C93D9 Fab
Method: single particle / : Windsor IW, Tong P, Gautam AK, Wesemann DR, Harrison SC

EMDB-24197: 
SARS-CoV-2 spike (6P) in complex with C81C10 Fab
Method: single particle / : Windsor IW, Tong P, Gautam AK, Wesemann DR, Harrison SC

EMDB-24198: 
SARS-CoV-2 spike (2P) in complex with C12A2 Fab
Method: single particle / : Bajic G, Windsor IW, Tong P, Gautam A, Wesemann DR, Harrison SC

PDB-7n62: 
SARS-CoV-2 Spike (2P) in complex with C12C9 Fab (NTD local reconstruction)
Method: single particle / : Windsor IW, Jenni S, Bajic G, Tong P, Gautam AK, Wesemann DR, Harrison SC

PDB-7n64: 
SARS-CoV-2 Spike (2P) in complex with G32R7 Fab (RBD and NTD local reconstruction)
Method: single particle / : Windsor IW, Jenni S, Tong P, Gautam AK, Wesemann DR, Harrison SC

EMDB-3944: 
BK polyomavirus + 20 mM GT1b oligosaccharide
Method: single particle / : Hurdiss DL, Ranson NA

EMDB-3945: 
BK polyomavirus + heparin
Method: single particle / : Hurdiss DL, Ranson NA

EMDB-3946: 
BK polyomavirus
Method: single particle / : Hurdiss DL, Ranson NA

EMDB-4212: 
BK polyomavirus + 5 mM dithiothreitol
Method: single particle / : Hurdiss DL, Ranson NA

PDB-6esb: 
BK polyomavirus + 20 mM GT1b oligosaccharide
Method: single particle / : Hurdiss DL, Ranson NA

EMDB-3600: 
Contracted sheath of a Pseudomonas aeruginosa type six secretion system consisting of TssB1 and TssC1
Method: helical / : Salih O, He S, Stach L, Macdonald JT, Planamente S, Manoli E, Scheres S, Filloux A, Freemont PS

PDB-5n8n: 
Contracted sheath of a Pseudomonas aeruginosa type six secretion system consisting of TssB1 and TssC1
Method: helical / : Salih O, He S, Stach L, Macdonald JT, Planamente S, Manoli E, Scheres S, Filloux A, Freemont PS

EMDB-3283: 
Cryo-EM structure of BK polyomavirus
Method: single particle / : Hurdiss DL, Morgan EL, Thompson RF, Prescott EL, Panou MM, Macdonald A, Ranson NA

EMDB-3284: 
Cryo-EM structure of BK polyomavirus VP1 virus-like particle
Method: single particle / : Hurdiss DL, Morgan EL, Thompson RF, Prescott EL, Panou MM, Macdonald A, Ranson NA

PDB-5fua: 
Cryo-EM of BK polyomavirus
Method: single particle / : Hurdiss DL, Morgan EL, Thompson RF, Prescott EL, Panou MM, Macdonald A, Ranson NA

EMDB-5494: 
Cryo-EM structure of short shafted adenovirus type 5 complexed with factor X
Method: single particle / : Doronin K, Flatt JW, Di Paolo NC, Khare R, Kalyuzhniy O, Acchione M, Sumida JP, Ohto U, Shimizu T, Akashi-Takamura S, Miyake K, MacDonald JW, Bammler TK, Beyer RP, Farin FM, Stewart PL, Shayakhmetov DM
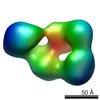
EMDB-1964: 
Structural and Functional Studies of LRP6 Ectodomain Reveal a Platform for Wnt Signaling
Method: single particle / : Chen S, Bubeck D, MacDonald BT, Liang WX, Mao JH, Malinauskas T, Llorca O, Aricescu AR, Siebold C, He X, Jones EY
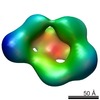
EMDB-1965: 
Structural and Functional Studies of LRP6 Ectodomain Reveal a Platform for Wnt Signaling
Method: single particle / : Chen S, Bubeck D, MacDonald BT, Liang WX, Mao JH, Malinauskas T, Llorca O, Aricescu AR, Siebold C, He X, Jones EY
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
