-Search query
-Search result
Showing 1 - 50 of 167 items for (author: moores & ca)
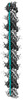
EMDB-19042: 
MAP7 MTBD (microtubule binding domain) decorated microtubule protofilament
Method: single particle / : Bangera M, Moores CA

EMDB-19043: 
MAP7 MTBD (microtubule binding domain) decorated Taxol stabilized microtubule (13pf) protofilament
Method: single particle / : Bangera M, Moores CA

EMDB-19044: 
MAP7 MTBD (microtubule binding domain) decorated Taxol stabilized microtubule (14pf) protofilament
Method: single particle / : Bangera M, Moores CA

PDB-8rc1: 
MAP7 MTBD (microtubule binding domain) decorated microtubule protofilament
Method: single particle / : Bangera M, Moores CA

EMDB-17557: 
Cryo-EM structure of cortactin-stabilized Arp2/3-complex nucleated actin branches-Local refined map on mother filament
Method: single particle / : Liu T, Moores CA

EMDB-17553: 
Cryo-EM structure of cortactin-stabilized Arp2/3 complex nucleated actin branches-Daughter filament consensus map
Method: single particle / : Liu T, Moores CA

EMDB-17554: 
Cryo-EM structure of cortactin-stabilized Arp2/3 nucleated actin branches-Local refined map on Arp2/3 complex
Method: single particle / : Liu T, Moores CA

EMDB-17555: 
Cryo-EM structure of cortactin-stabilized Arp2/3-complex nucleated actin branches-Local refined map on the daughter filament and cortactin density
Method: single particle / : Liu T, Moores CA

EMDB-17556: 
Cryo-EM structure of cortactin-stabilized Arp2/3-complex nucleated actin branches-Local refined map on capping protein
Method: single particle / : Liu T, Moores CA

EMDB-17558: 
Cryo-EM structure of cortactin stabilized Arp2/3-complex nucleated actin branches
Method: single particle / : Liu T, Moores CA

PDB-8p94: 
Cryo-EM structure of cortactin stabilized Arp2/3-complex nucleated actin branches
Method: single particle / : Liu T, Moores CA
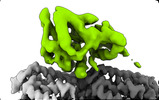
EMDB-14460: 
P. berghei kinesin-8B motor domain in AMPPNP state bound to tubulin dimer
Method: single particle / : Liu T, Shilliday F, Cook AD, Moores CA

PDB-7z2b: 
P. berghei kinesin-8B motor domain in AMPPNP state bound to tubulin dimer
Method: single particle / : Liu T, Shilliday F, Cook AD, Moores CA

EMDB-14459: 
P. berghei kinesin-8B motor domain in no nucleotide state bound to tubulin dimer
Method: single particle / : Liu T, Shilliday F, Cook AD, Moores CA
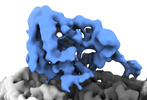
EMDB-14461: 
P. falciparum kinesin-8B motor domain in no nucleotide bound to tubulin dimer
Method: single particle / : Liu T, Shilliday F, Cook AD, Moores CA

PDB-7z2a: 
P. berghei kinesin-8B motor domain in no nucleotide state bound to tubulin dimer
Method: single particle / : Liu T, Shilliday F, Cook AD, Moores CA

PDB-7z2c: 
P. falciparum kinesin-8B motor domain in no nucleotide bound to tubulin dimer
Method: single particle / : Liu T, Shilliday F, Cook AD, Moores CA

EMDB-14386: 
In situ subtomogram average of long-repeat F-actin from neuronal growth cones
Method: subtomogram averaging / : Atherton J, Stouffer M, Francis F, Moores CA
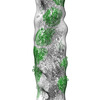
EMDB-14395: 
In situ subtomogram average of short-repeat F-actin from neuronal growth cones
Method: subtomogram averaging / : Atherton J, Stouffer M, Francis F, Moores CA

EMDB-14396: 
Tomogram of wild-type mouse neuronal growth cone (T-zone), 4 x binned, processed.
Method: electron tomography / : Atherton J, Stouffer M, Francis F, Moores CA

EMDB-14397: 
Tomogram of wild-type mouse neuronal growth cone (P-zone), 4 x binned, processed.
Method: electron tomography / : Atherton J, Stouffer M, Francis F, Moores CA

EMDB-14400: 
Tomogram of wild-type mouse neuronal growth cone (C-zone), 4 x binned, processed.
Method: electron tomography / : Atherton J, Stouffer M, Francis F, Moores CA

EMDB-14401: 
Tomogram of doublecortin knock-out mouse neuronal growth cone (C-zone), 4 x binned, processed.
Method: electron tomography / : Atherton J, Stouffer M, Francis F, Moores CA

EMDB-14402: 
Tomogram of doublecortin knock-out mouse neuronal growth cone (P-zone), 4 x binned, processed.
Method: electron tomography / : Atherton J, Stouffer M, Francis F, Moores CA
EMDB-14414: 
Tomogram of doublecortin knock-out mouse neuronal growth cone (T-zone), 4 x binned, processed.
Method: electron tomography / : Atherton J, Stouffer M, Francis F, Moores CA

EMDB-14416: 
Tomogram of wild-type mouse neuronal growth cone (P-zone), 2 x binned, processed.
Method: electron tomography / : Atherton J, Stouffer M, Francis F, Moores CA

EMDB-12257: 
Plasmodium falciparum kinesin-5 motor domain without nucleotide, complexed with 14 protofilament microtubule.
Method: helical / : Cook AD, Roberts A, Atherton J, Tewari R, Topf M, Moores CA

EMDB-12258: 
Plasmodium falciparum kinesin-5 motor domain bound to AMPPNP, complexed with 14 protofilament microtubule.
Method: helical / : Cook AD, Roberts A, Atherton J, Tewari R, Topf M, Moores CA

PDB-7nb8: 
Plasmodium falciparum kinesin-5 motor domain without nucleotide, complexed with 14 protofilament microtubule.
Method: helical / : Cook AD, Roberts A, Atherton J, Tewari R, Topf M, Moores CA

PDB-7nba: 
Plasmodium falciparum kinesin-5 motor domain bound to AMPPNP, complexed with 14 protofilament microtubule.
Method: helical / : Cook AD, Roberts A, Atherton J, Tewari R, Topf M, Moores CA

EMDB-11338: 
Kinesin binding protein (KBP)
Method: single particle / : Atherton J, Hummel JJA, Olieric N, Locke J, Pena A, Rosenfeld SS, Steinmetz MO, Hoogenraad CC, Moores CA

EMDB-11339: 
Kinesin binding protein complexed with Kif15 motor domain
Method: single particle / : Atherton J, Hummel JJA, Olieric N, Locke J, Pena A, Rosenfeld SS, Steinmetz MO, Hoogenraad CC, Moores CA

EMDB-11340: 
Microtubule complexed with Kif15 motor domain. Symmetrised asymmetric unit
Method: single particle / : Atherton J, Hummel JJA, Olieric N, Locke J, Pena A, Rosenfeld SS, Steinmetz MO, Hoogenraad CC, Moores CA

PDB-6zpg: 
Kinesin binding protein (KBP)
Method: single particle / : Atherton J, Hummel JJA, Olieric N, Locke J, Pena A, Rosenfeld SS, Steinmetz MO, Hoogenraad CC, Moores CA

PDB-6zph: 
Kinesin binding protein complexed with Kif15 motor domain
Method: single particle / : Atherton J, Hummel JJA, Olieric N, Locke J, Pena A, Rosenfeld SS, Steinmetz MO, Hoogenraad CC, Moores CA

PDB-6zpi: 
Microtubule complexed with Kif15 motor domain. Symmetrised asymmetric unit
Method: single particle / : Atherton J, Hummel JJA, Olieric N, Locke J, Pena A, Rosenfeld SS, Steinmetz MO, Hoogenraad CC, Moores CA

EMDB-10959: 
Cryo-EM structure of the ARP2/3 1B5CL isoform complex.
Method: single particle / : von Loeffelholz O, Moores C, Purkiss A

EMDB-10960: 
Cryo-EM structure of the ARP2/3 1A5C isoform complex.
Method: single particle / : von Loeffelholz O, Moores C, Purkiss A

PDB-6yw6: 
Cryo-EM structure of the ARP2/3 1B5CL isoform complex.
Method: single particle / : von Loeffelholz O, Moores C, Purkiss A

PDB-6yw7: 
Cryo-EM structure of the ARP2/3 1A5C isoform complex.
Method: single particle / : von Loeffelholz O, Moores C, Purkiss A

EMDB-4858: 
Cryo-EM structure of the N-terminal DC repeat (NDC) of human doublecortin (DCX) bound to 13-protofilament GDP-microtubule
Method: single particle / : Manka SW

EMDB-4861: 
Cryo-EM structure of the C-terminal DC repeat (CDC) of human doublecortin (DCX) bound to 13-protofilament GDP.Pi-microtubule
Method: single particle / : Manka SW

EMDB-4863: 
Cryo-EM structure of the N-terminal DC repeat (NDC) of NDC-NDC chimera (human sequence) bound to 14-protofilament GDP-microtubule
Method: single particle / : Manka SW

PDB-6rev: 
Cryo-EM structure of the N-terminal DC repeat (NDC) of human doublecortin (DCX) bound to 13-protofilament GDP-microtubule
Method: single particle / : Manka SW

PDB-6rf2: 
Cryo-EM structure of the C-terminal DC repeat (CDC) of human doublecortin (DCX) bound to 13-protofilament GDP.Pi-microtubule
Method: single particle / : Manka SW

PDB-6rfd: 
Cryo-EM structure of the N-terminal DC repeat (NDC) of NDC-NDC chimera (human sequence) bound to 14-protofilament GDP-microtubule
Method: single particle / : Manka SW

EMDB-10421: 
Human kinesin-5 motor domain in the GSK-1 state bound to microtubules
Method: helical / : Pena A, Sweeney A, Cook AD, Moores CA, Topf M

PDB-6ta3: 
Human kinesin-5 motor domain in the GSK-1 state bound to microtubules (Conformation 1)
Method: helical / : Pena A, Sweeney A, Cook AD, Moores CA, Topf M

EMDB-4862: 
Cryo-EM structure of the N-terminal DC repeat (NDC) of NDC-NDC chimera (human sequence) bound to 13-protofilament GDP-microtubule
Method: single particle / : Manka SW

PDB-6rf8: 
Cryo-EM structure of the N-terminal DC repeat (NDC) of NDC-NDC chimera (human sequence) bound to 13-protofilament GDP-microtubule
Method: single particle / : Manka SW
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
