-Search query
-Search result
Showing all 47 items for (author: m. & a. & williams)

PDB-8quc: 
Cryo-EM Structure of Human Kv3.1 in Complex with Modulator AUT1
Method: single particle / : Chi G, Mckinley G, Marsden B, Pike ACW, Ye M, Brooke LM, Bakshi S, Pilati N, Marasco A, Gunthorpe M, Alvaro G, Large C, Lakshminaraya B, Williams E, Sauer DB

PDB-8qud: 
Cryo-EM Structure of Human Kv3.1 in Complex with Modulator AUT5
Method: single particle / : Chi G, Mckinley G, Marsden B, Pike ACW, Ye M, Brooke LM, Bakshi S, Lakshminarayana B, Pilati N, Marasco A, Gunthorpe M, Alvaro GS, Large CH, Williams E, Sauer DB

PDB-8c54: 
Cryo-EM structure of NADH bound SLA dehydrogenase RlGabD from Rhizobium leguminosarum bv. trifolii SRD1565
Method: single particle / : Sharma M, Meek RW, Armstrong Z, Blaza JN, Alhifthi A, Li J, Goddard-Borger ED, Williams SJ, Davies GJ

PDB-8afz: 
Architecture of the ESCPE-1 membrane coat
Method: subtomogram averaging / : Lopez-Robles C, Scaramuzza S, Astorga-Simon E, Ishida M, Williamsom CD, Banos-Mateos S, Gil-Carton D, Romero M, Vidaurrazaga A, Fernandez-Recio J, Rojas AL, Bonifacino JS, Castano-Diez D, Hierro A

PDB-8a1r: 
cryo-EM structure of thioredoxin glutathione reductase in complex with a non-competitive inhibitor
Method: single particle / : Ardini M, Angelucci F, Fata F, Gabriele F, Effantin G, Ling W, Williams DL, Petukhova VZ, Petukhov PA

PDB-8f2r: 
Human CCC complex
Method: single particle / : Healy MD, McNally KE, Butkovic R, Chilton M, Kato K, Sacharz J, McConville C, Moody ERR, Shaw S, Planelles-Herrero VJ, Kadapalakere SY, Ross J, Borucu U, Palmer CS, Chen K, Croll TI, Hall RJ, Caruana NJ, Ghai R, Nguyen THD, Heesom KJ, Saitoh S, Berger I, Berger-Schaffitzel C, Williams TA, Stroud DA, Derivery E, Collins BM, Cullen PJ

PDB-8f2u: 
Human CCC complex
Method: single particle / : Healy MD, McNally KE, Butkovic R, Chilton M, Kato K, Sacharz J, McConville C, Moody ERR, Shaw S, Planelles-Herrero VJ, Kadapalakere SY, Ross J, Borucu U, Palmer CS, Chen K, Croll TI, Hall RJ, Caruana NJ, Ghai R, Nguyen THD, Heesom KJ, Saitoh S, Berger I, Berger-Schaffitzel C, Williams TA, Stroud DA, Derivery E, Collins BM, Cullen PJ

PDB-7tn9: 
Structure of the Inmazeb cocktail and resistance to escape against Ebola virus
Method: single particle / : Rayaprolu V, Fulton B, Rafique A, Arturo E, Williams D, Hariharan C, Callaway H, Parvate A, Schendel SL, Parekh D, Hui S, Shaffer K, Pascal KE, Wloga E, Giordano S, Copin R, Franklin M, Boytz RM, Donahue C, Davey R, Baum A, Kyratsous CA, Saphire EO

PDB-8bc2: 
Ligand-Free Structure of the decameric sulfofructose transaldolase BmSF-TAL
Method: single particle / : Snow AJD, Sharma M, Blaza J, Davies GJ

PDB-8bc3: 
Cryo-EM Structure of a BmSF-TAL - Sulfofructose Schiff Base Complex
Method: single particle / : Snow AJD, Sharma M, Blaza J, Davies GJ

PDB-8bc4: 
Cryo-EM Structure of a BmSF-TAL - Sulfofructose Schiff Base Complex in symmetry group C1
Method: single particle / : Snow AJD, Sharma M, Blaza J, Davies GJ
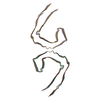
PDB-7upe: 
Tau Paired Helical Filament from Alzheimer's Disease not incubated with EGCG
Method: helical / : Seidler PM, Murray KA, Boyer DR, Ge P, Sawaya MR, Eisenberg DS
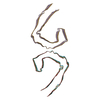
PDB-7upf: 
Tau Paired Helical Filament from Alzheimer's Disease incubated 1 hr. with EGCG
Method: helical / : Seidler PM, Murray KA, Boyer DR, Ge P, Sawaya MR, Eisenberg DS
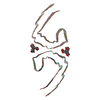
PDB-7upg: 
Tau Paired Helical Filament from Alzheimer's Disease incubated with EGCG for 3 hours
Method: helical / : Seidler PM, Murray KA, Boyer DR, Ge P, Sawaya MR, Eisenberg DS

PDB-7t8b: 
Octameric Human Twinkle Helicase Clinical Variant W315L
Method: single particle / : Riccio AA, Bouvette J, Krahn J, Borgnia MJ, Copeland WC

PDB-7t8c: 
Heptameric Human Twinkle Helicase Clinical Variant W315L
Method: single particle / : Riccio AA, Bouvette J, Krahn J, Borgnia MJ, Copeland WC

PDB-7owg: 
human DEPTOR in a complex with mutant human mTORC1 A1459P
Method: single particle / : Heimhalt M, Berndt A, Wagstaff J, Anandapadamanaban M, Perisic O, Maslen S, McLaughlin S, Yu WH, Masson GR, Boland A, Ni X, Yamashita K, Murshudov GN, Skehel M, Freund SM, Williams RL

PDB-6zvr: 
C11 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Method: single particle / : Liu JW, Tassinari M, Souza DP, Naskar S, Noel JK, Bohuszewicz O, Buck M, Williams TA, Baum B, Low HH

PDB-6zvs: 
C12 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Method: single particle / : Liu J, Tassinari M, Souza DP, Naskar S, Noel JK, Bohuszewicz O, Buck M, Williams TA, Baum B, Low HH

PDB-6zvt: 
C13 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Method: single particle / : Liu JW, Tassinari M, Souza DP, Naskar S, Noel JK, Bohuszewicz O, Buck M, Williams TA, Baum B, Low HH

PDB-6zw4: 
C14 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Method: single particle / : Liu JW, Tassinari M, Souza DP, Naskar S, Noel JK, Bohuszewicz O, Buck M, Williams TA, Baum B, Low HH

PDB-6zw5: 
C15 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Method: single particle / : Liu JW, Tassinari M, Souza DP, Naskar S, Noel JK, Bohuszewicz O, Buck M, Williams TA, Baum B, Low HH

PDB-6zw6: 
C16 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Method: single particle / : Liu J, Tassinari M, Souza DP, Naskar S, Noel JK, Bohuszewicz O, Buck M, Williams TA, Baum B, Low HH

PDB-6zw7: 
C17 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Method: single particle / : Liu J, Tassinari M, Souza DP, Naskar S, Noel JK, Bohuszewicz O, Buck M, Williams TA, Baum B, Low HH

PDB-6wiv: 
Structure of human GABA(B) receptor in an inactive state
Method: single particle / : Park J, Fu Z, Frangaj A, Liu J, Mosyak L, Shen T, Slavkovich VN, Ray KM, Taura J, Cao B, Geng Y, Zuo H, Kou Y, Grassucci R, Chen S, Liu Z, Lin X, Williams JP, Rice WJ, Eng ET, Huang RK, Soni RK, Kloss B, Yu Z, Javitch JA, Hendrickson WA, Slesinger PA, Quick M, Graziano J, Yu H, Fiehn O, Clarke OB, Frank J, Fan QR
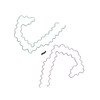
PDB-6vi3: 
Straight Filament from Alzheimer's Disease Human Brain Tissue
Method: helical / : Arakhamia T, Lee CE, Carlomagno Y, Duong DM, Kundinger SR, Wang K, Williams D, DeTure M, Dickson DW, Cook CN, Seyfried NT, Petrucelli L, Fitzpatrick AWP

PDB-6pdt: 
cryoEM structure of yeast glucokinase filament
Method: helical / : Lynch EM, Dosey AM, Farrell DP, Stoddard PR, Kollman JM
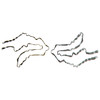
PDB-6vh7: 
Doublet Tau Fibril from Corticobasal Degeneration Human Brain Tissue
Method: helical / : Arakhamia T, Lee CE, Carlomagno Y, Duong DM, Kundinger SR, Wang K, Williams D, DeTure M, Dickson DW, Cook CN, Seyfried NT, Petrucelli L, Fitzpatrick AWP
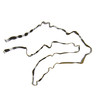
PDB-6vha: 
Singlet Tau Fibril from Corticobasal Degeneration Human Brain Tissue
Method: helical / : Arakhamia T, Lee CE, Carlomagno Y, Duong DM, Kundinger SR, Wang K, Williams D, DeTure M, Dickson DW, Cook CN, Seyfried NT, Petrucelli L, Fitzpatrick AWP

PDB-6vhl: 
Paired Helical Filament from Alzheimer's Disease Human Brain Tissue
Method: helical / : Arakhamia T, Lee CE, Carlomagno Y, Duong DM, Kundinger SR, Wang K, Williams D, DeTure M, Dickson DW, Cook CN, Seyfried NT, Petrucelli L, Fitzpatrick AWP

PDB-6pnj: 
Structure of Photosystem I Acclimated to Far-red Light
Method: single particle / : Gisriel CJ, Shen G, Kurashov V, Ho M, Zhang S, Williams D, Golbeck JH, Fromme P, Bryant DA

PDB-6tte: 
Beta-galactosidase in complex with PETG
Method: single particle / : Saur M, Hartshorn MJ, Dong J, Reeks J, Bunkoczi G, Jhoti H, Williams PA

PDB-6ttf: 
PKM2 in complex with Compound 5
Method: single particle / : Saur M, Hartshorn MJ, Dong J, Reeks J, Bunkoczi G, Jhoti H, Williams PA

PDB-6tth: 
PKM2 in complex with L-threonine
Method: single particle / : Saur M, Hartshorn MJ, Dong J, Reeks J, Bunkoczi G, Jhoti H, Williams PA

PDB-6tti: 
PKM2 in complex with Compound 6
Method: single particle / : Saur M, Hartshorn MJ, Dong J, Reeks J, Bunkoczi G, Jhoti H, Williams PA

PDB-6ttq: 
PKM2 in complex with Compound 10
Method: single particle / : Saur M, Hartshorn MJ, Dong J, Reeks J, Bunkoczi G, Jhoti H, Williams PA

PDB-6tsh: 
Beta-galactosidase in complex with deoxygalacto-nojirimycin
Method: single particle / : Saur M, Hartshorn MJ, Dong J, Reeks J, Bunkoczi G, Jhoti H, Williams PA

PDB-6tsk: 
Beta-galactosidase in complex with L-ribose
Method: single particle / : Saur M, Hartshorn MJ, Dong J, Reeks J, Bunkoczi G, Jhoti H, Williams PA

PDB-6s8f: 
Structure of nucleotide-bound Tel1/ATM
Method: single particle / : Yates LA, Williams RM, Ayala R, Zhang X

PDB-6sb0: 
cryo-EM structure of mTORC1 bound to PRAS40-fused active RagA/C GTPases
Method: single particle / : Anandapadamanaban M, Berndt A, Masson GR, Perisic O, Williams RL

PDB-6sb2: 
cryo-EM structure of mTORC1 bound to active RagA/C GTPases
Method: single particle / : Anandapadamanaban M, Berndt A, Masson GR, Perisic O, Williams RL

PDB-6of2: 
Precursor ribosomal RNA processing complex, State 2.
Method: single particle / : Pillon MC, Hsu AL, Krahn JM, Williams JG, Goslen KH, Sobhany M, Borgnia MJ, Stanley RE

PDB-6of3: 
Precursor ribosomal RNA processing complex, State 1.
Method: single particle / : Pillon MC, Hsu AL, Krahn JM, Williams JG, Goslen KH, Sobhany M, Borgnia MJ, Stanley RE

PDB-6of4: 
Precursor ribosomal RNA processing complex, apo-state.
Method: single particle / : Pillon MC, Hsu AL, Krahn JM, Williams JG, Goslen KH, Sobhany M, Borgnia MJ, Stanley RE

PDB-5np0: 
Closed dimer of human ATM (Ataxia telangiectasia mutated)
Method: single particle / : Baretic D, Pollard HK, Fisher DI, Johnson CM, Santhanam B, Truman CM, Kouba T, Fersht AR, Phillips C, Williams RL

PDB-5np1: 
Open protomer of human ATM (Ataxia telangiectasia mutated)
Method: single particle / : Baretic D, Pollard HK, Fisher DI, Johnson CM, Santhanam B, Truman CM, Kouba T, Fersht AR, Phillips C, Williams RL

PDB-5fvm: 
Cryo electron microscopy of a complex of Tor and Lst8
Method: single particle / : Baretic D, Berndt A, Ohashi Y, Johnson CM, Williams RL
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
