-Search query
-Search result
Showing 1 - 50 of 110 items for (author: liu & yf)
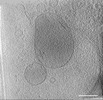
EMDB-39012: 
Representative tomogram of primary glioblastoma stem cell with circular inter-mitochondrial junctions.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39015: 
Representative tomogram of microglia cell with nanotunnel-like structures resembling mitochondrial fission.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT
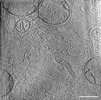
EMDB-39019: 
Representative tomogram of glioblastoma cell with nanotunnel-like structure and inter-mitochondrial junction.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39021: 
Representative tomogram of normal human astrocyte with nanotunnel-like structure which is an extension of the mitochondrial outer membrane.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39023: 
Representative tomogram of primary glioblastoma differentiated cell with parallel inter-mitochondrial junction.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39024: 
Representative tomogram of primary glioblastoma stem cell with clustered mitochondria bearing various long-short axis ratios.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-33990: 
Cryo-EM structure of EBV gHgL-gp42 in complex with mAbs 3E8 and 5E3 (localized refinement)
Method: single particle / : Liu L, Sun H, Jiang Y, Hong J, Zheng Q, Li S, Chen Y, Xia N
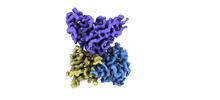
EMDB-33992: 
Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 10E4 (localized refinement)
Method: single particle / : Liu L, Sun H, Jiang Y, Hong J, Zheng Q, Li S, Chen Y, Xia N
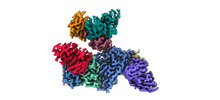
EMDB-33993: 
Cryo-EM density map of EBV gHgL-gp42 in complex with four mAbs 5E3, 3E8, 6H2 and 10E4
Method: single particle / : Liu L, Sun H, Jiang Y, Liu X, Zhao D, Zheng Q, Li S, Chen Y, Xia N
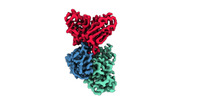
EMDB-33994: 
Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 6H2 (localized refinement)
Method: single particle / : Liu L, Sun H, Jiang Y, Hong J, Zheng Q, Li S, Chen Y, Xia N

EMDB-35758: 
Cryo-EM structure of mouse BIRC6, N-terminal section optimized
Method: single particle / : Liu S, Jiang T, Bu F, Zhao J, Wang G, Li N, Gao N, Qiu X

EMDB-35759: 
Cryo-EM structure of mouse BIRC6, Global map
Method: single particle / : Liu S, Jiang T, Bu F, Zhao J, Wang G, Li N, Gao N, Qiu X
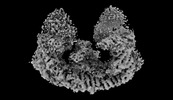
EMDB-35760: 
Cryo-EM structure of mouse BIRC6, with endogenous Smac binding
Method: single particle / : Liu S, Jiang T, Bu F, Zhao J, Wang G, Li N, Gao N, Qiu X

EMDB-38461: 
Cryo-EM structure of mouse BIRC6, Core region
Method: single particle / : Liu S, Jiang T, Bu F, Zhao J, Wang G, Li N, Gao N, Qiu X

EMDB-38462: 
Cryo-EM structure of mouse BIRC6, Half map of the core region
Method: single particle / : Liu S, Jiang T, Bu F, Zhao J, Wang G, Li N, Gao N, Qiu X

EMDB-38464: 
Cryo-EM structure of mouse BIRC6, Composite map
Method: single particle / : Liu S, Jiang T, Bu F, Zhao J, Wang G, Li N, Gao N, Qiu X

EMDB-29943: 
Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein
Method: single particle / : Shenming H, Mengyao X, Lei L, Yang D, Sheng C, Jinpeng S

EMDB-29944: 
Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein
Method: single particle / : Shenming H, Mengyao X, Lei L, Jiangqian M, Sheng C, Jinpeng S

EMDB-29945: 
Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein
Method: single particle / : Shenming H, Mengyao X, Lei L, Jianqiang M, Jinpeng S

EMDB-29946: 
Cryo-EM Structure of the Prostaglandin E2 Receptor 3 Coupled to G Protein
Method: single particle / : Shenming H, Mengyao X, Lei L, Yang D, Shiyi G, Jinpeng S

EMDB-29935: 
Cryo-EM Structure of the Prostaglandin E Receptor EP4 Coupled to G Protein
Method: single particle / : Huang SM, Xiong MY, Liu L, Mu J, Sheng C, Sun J

EMDB-29940: 
Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein
Method: single particle / : Huang SM, Xiong MY, Liu L, Mu J, Sheng C, Sun J

EMDB-33989: 
Cryo-EM structure of the N-terminal domain of hMCM8/9 and HROB
Method: single particle / : Zheng JF, Weng ZF, Liu YF
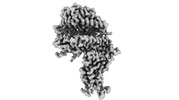
EMDB-33841: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.2.12.1 RBD in complex with human ACE2 (local refinement)
Method: single particle / : Zhao ZN, Xie YF, Qi JX, Gao GF
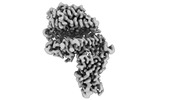
EMDB-33870: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with human ACE2 (local refinement)
Method: single particle / : Zhao ZN, Xie YF, Qi JX, Gao GF
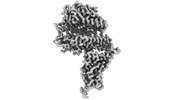
EMDB-34120: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with golden hamster ACE2 (local refinement)
Method: single particle / : Zhao ZN, Xie YF, Chai Y, Qi JX, Gao GF

EMDB-34138: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with mouse ACE2 (local refinement)
Method: single particle / : Zhao ZN, Xie YF, Chai Y, Qi JX, Gao GF

EMDB-34217: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with rat ACE2 (local refinement)
Method: single particle / : Zhao ZN, Xie YF, Chai Y, Qi JX, Gao GF

EMDB-34409: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.4/5 RBD in complex with human ACE2 (local refinement)
Method: single particle / : Zhao ZN, Xie YF, Qi JX, Gao GF

EMDB-34494: 
Cryo-EM map of SARS-CoV-2 Omicron BA.2 spike trimer in complex with human ACE2 (three-RBD-up conformation)
Method: single particle / : Zhao ZN, Xie YF, Qi JX, Gao GF

EMDB-34498: 
Cryo-EM map of SARS-CoV-2 Omicron BA.2.12.1 spike trimer in complex with human ACE2 (three-RBD-up conformation)
Method: single particle / : Zhao ZN, Xie YF, Qi JX, Gao GF

EMDB-34499: 
Cryo-EM map of SARS-CoV-2 Omicron BA.2 spike protein in complex with mouse ACE2
Method: single particle / : Zhao ZN, Xie YF, Chai Y, Qi JX, Gao GF

EMDB-34506: 
Cryo-EM map of SARS-CoV-2 Omicron BA.2 spike trimer in complex with rat ACE2
Method: single particle / : Zhao ZN, Xie YF, Chai Y, Qi JX, Gao GF

EMDB-34509: 
Cryo-EM map of SARS-CoV-2 Omicron BA.4/5 (N658S) spike trimer in complex with human ACE2 (three-RBD-up conformation)
Method: single particle / : Zhao ZN, Xie YF, Qi JX, Gao GF
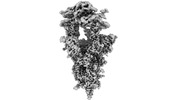
EMDB-34510: 
Cryo-EM map of SARS-CoV-2 Omicron BA.2 spike trimer in complex with golden hamster ACE2
Method: single particle / : Zhao ZN, Xie YF, Chai Y, Qi JX, Gao GF

EMDB-35683: 
Dopamine Receptor D1R-Gs-Rotigotine complex
Method: single particle / : Xu P, Huang S, Zhuang Y, Mao C, Zhang Y, Wang Y, Li H, Jiang Y, Xu HE

EMDB-35686: 
Dopamine Receptor D4R-Gi-Rotigotine complex
Method: single particle / : Xu P, Huang S, Zhuang Y, Mao C, Zhang Y, Wang Y, Li H, Jiang Y, Xu HE

EMDB-35684: 
Dopamine Receptor D2R-Gi-Rotigotine complex
Method: single particle / : Xu P, Huang S, Zhuang Y, Mao C, Zhang Y, Wang Y, Li H, Jiang Y, Xu HE

EMDB-35685: 
Dopamine Receptor D3R-Gi-Rotigotine complex
Method: single particle / : Xu P, Huang S, Zhuang Y, Mao C, Zhang Y, Wang Y, Li H, Jiang Y, Xu HE

EMDB-35687: 
Dopamine Receptor D5R-Gs-Rotigotine complex
Method: single particle / : Xu P, Huang S, Zhuang Y, Mao C, Zhang Y, Wang Y, Li H, Jiang Y, Xu HE

EMDB-35705: 
Cryo-EM structure of the DMCHA-bound mTAAR9-Gs complex
Method: single particle / : Sun JP, Li Q, Yang F, Xu YF, Guo LL, Lian S, Zhang MH, Rong NK

EMDB-35761: 
Cryo-EM structure of the PEA-bound mTAAR9-Golf complex
Method: single particle / : Sun JP, Li Q, Yang F, Xu YF, Guo LL, Lian S, Zhang MH, Rong NK

EMDB-35762: 
Cryo-EM structure of the SPE-bound mTAAR9-Gs complex
Method: single particle / : Sun JP, Li Q, Yang F, Xu YF, Guo LL, Lian S, Zhang MH, Rong NK

EMDB-35763: 
Cryo-EM structure of the PEA-bound mTAAR9-Gs complex
Method: single particle / : Sun JP, Li Q, Yang F, Xu YF, Guo LL, Lian S, Zhang MH, Rong NK

EMDB-35764: 
Cryo-EM structure of the CAD-bound mTAAR9-Gs complex
Method: single particle / : Sun JP, Li Q, Yang F, Xu YF, Guo LL, Lian S, Zhang MH, Rong NK

EMDB-35765: 
Cryo-EM structure of the SPE-mTAAR9 complex
Method: single particle / : Sun JP, Li Q, Yang F, Xu YF, Guo LL, Lian S, Zhang MH, Rong NK

EMDB-35771: 
Cryo-EM structure of the PEA-bound mTAAR9 complex
Method: single particle / : Sun JP, Li Q, Yang F, Xu YF, Guo LL, Lian S, Zhang MH, Rong NK

EMDB-32346: 
Cryo-EM structure of gMCM8/9 helicase
Method: single particle / : Zheng JF, Weng ZF, Liu YF

EMDB-32356: 
Computational design of a potent Epstein-Barr virus fusion protein gB nanoparticle vaccine
Method: subtomogram averaging / : Sun C, Wang AJ, Fang XY, Gao YZ, Liu Z, Zeng MS

EMDB-33803: 
Cryo-EM structure of human sodium-chloride cotransporter
Method: single particle / : Nan J, Yang XM, Shan ZY, Yuan YF, Zhang YQ
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
