-Search query
-Search result
Showing all 32 items for (author: liu & xq)

EMDB-35706: 
Cryo-EM structure of GIPR splice variant 1 (SV1) in complex with Gs protein
Method: single particle / : Zhao FH, Hang KN, Zhou QT, Shao LJ, Li H, Li WZ, Lin S, Dai AT, Cai XQ, Liu YY, Xu YN, Feng WB, Yang DH, Wang MW

EMDB-35707: 
Cryo-EM structure of GIPR splice variant 2 (SV2) in complex with Gs protein
Method: single particle / : Zhao FH, Hang KN, Zhou QT, Shao LJ, Li H, Li WZ, Lin S, Dai AT, Cai XQ, Liu YY, Xu YN, Feng WB, Yang DH, Wang MW

EMDB-35601: 
Cryo-EM structure of the alpha-MSH-bound human melanocortin receptor 5 (MC5R)-Gs complex
Method: single particle / : Feng WB, Zhou QT, Chen XY, Dai AT, Cai XQ, Liu X, Zhao FH, Chen Y, Ye CY, Xu YN, Cong ZT, Li H, Lin S, Yang DH, Wang MW

EMDB-35615: 
Cryo-EM structure of the gamma-MSH-bound human melanocortin receptor 3 (MC3R)-Gs complex
Method: single particle / : Feng WB, Zhou QT, Chen XY, Dai AT, Cai XQ, Liu X, Zhao FH, Chen Y, Ye CY, Xu YN, Cong ZT, Li H, Lin S

EMDB-35616: 
Cryo-EM structure of the PG-901-bound human melanocortin receptor 5 (MC5R)-Gs complex
Method: single particle / : Feng WB, Zhou QT, Chen XY, Dai AT, Cai XQ, Liu X, Zhao FH, Chen Y, Ye CY, Xu YN, Cong ZT, Li H, Lin S, Yang DH, Wang MW

EMDB-33871: 
Cryo-EM structure of the INSL5-bound human relaxin family peptidereceptor 4 (RXFP4)-Gi complex
Method: single particle / : Chen Y, Zhou QT, Wang J, Xu YW, Wang Y, Yan JH, Wang YB, Zhu Q, Zhao FH, Li CH, Chen CW, Cai XQ, Bathgate RAD, Shen C, Liu H, Xu HE, Yang DH, Wang MW

EMDB-33888: 
Cryo-EM structure of the compound 4-bound human relaxin family peptide receptor 4 (RXFP4)-Gi complex
Method: single particle / : Chen Y, Zhou QT, Wang J, Xu YW, Wang Y, Yan JH, Wang YB, Zhu Q, Zhao FH, Li CH, Chen CW, Cai XQ, Bathgate RAD, Shen C, Liu H, Xu HE, Yang DH, Wang MW

EMDB-33889: 
Cryo-EM structure of the DC591053-bound human relaxin family peptide receptor 4 (RXFP4)-Gi complex
Method: single particle / : Chen Y, Zhou QT, Wang J, Xu YW, Wang Y, Yan JH, Wang YB, Zhu Q, Zhao FH, Li CH, Chen CW, Cai XQ, Bathgate RAD, Shen C, Xu HE, Yang DH, Liu H, Wang MW

EMDB-31824: 
Cryo-EM structure of the SV1-Gs complex.
Method: single particle / : Cong ZT, Zhou FL, Zhang C, Zou XY, Zhang HB, Wang YZ, Zhou QT, Cai XQ, Liu QF, Li J, Shao LJ, Mao CY, Wang X, Wu JH, Xia T, Zhao LH, Jiang HL, Zhang Y, Xu HE, Cheng X, Yang DH, Wang MW

EMDB-30816: 
Cryo-EM structure of SARS-CoV2 RBD-ACE2 complex
Method: single particle / : Wang J, Lan J, Wang XQ, Wang HW

EMDB-31825: 
Cryo-EM structure of the GHRH-bound human GHRHR splice variant 1 complex
Method: single particle / : Cong ZT, Zhou FL, Zhang C, Zou XY, Zhang HB, Wang YZ, Zhou QT, Cai XQ, Liu QF, Li J, Shao LJ, Mao CY, Wang X, Wu JH, Xia T, Zhao LH, Jiang HL, Zhang Y, Xu HE, Chen X, Yang DH, Wang MW

EMDB-31387: 
Cryo-EM structure of an activated Cholecystokinin A receptor (CCKAR)-Gi complex
Method: single particle / : Liu QF, Yang DH, Zhuang YW, Croll TI, Cai XQ, Duan J, Dai AT, Yin WC, Ye CY, Zhou FL, Wu BL, Zhao Q, Xu HE, Wang MW, Jiang Y

EMDB-31388: 
Cryo-EM structure of an activated Cholecystokinin A receptor (CCKAR)-Gs complex
Method: single particle / : Liu QF, Yang DH, Zhuang YW, Croll TI, Cai XQ, Duan J, Dai AT, Yin WC, Ye CY, Zhou FL, Wu BL, Zhao Q, Xu HE, Wang MW, Jiang Y

EMDB-31389: 
Cryo-EM structure of an activated Cholecystokinin A receptor (CCKAR)-Gq complex
Method: single particle / : Liu QF, Yang DH, Zhuang YW, Croll TI, Cai XQ, Duan J, Dai AT, Yin WC, Ye CY, Zhou FL, Wu BL, Zhao Q, Xu HE, Wang MW, Jiang Y

EMDB-30529: 
S protein of SARS-CoV-2 in complex bound with P5A-3C12_1B
Method: single particle / : Yan RH, Wang RK, Ju B, Yu JF, Zhang YY, Liu N, Wang HW, Wang XQ, Zhang LQ, Zhou Q

EMDB-30530: 
S protein of SARS-CoV-2 in complex bound with P5A-3A1
Method: single particle / : Yan RH, Wang RK, Yu JF, Zhang YY, Liu N, Wang HW, Wang XQ, Zhang LQ, Zhou Q

EMDB-30531: 
S protein of SARS-CoV-2 in complex bound with P5A-3C12_2B
Method: single particle / : Yan RH, Wang RK, Ju B, Yu JF, Zhang YY, Liu N, Wang HW, Wang XQ, Zhang LQ, Zhou Q

EMDB-30306: 
Structural basis for cross-species recognition of COVID-19 virus spike receptor binding domain to bat ACE2
Method: single particle / : Liu KF, Wang J, Tan SG, Niu S, Wu LL, Zhang YF, Pan XQ, Meng YM, Chen Q, Wang QH, Wang HW, Qi JX, Gao GF

EMDB-20470: 
Cryo-EM structure of human cannabinoid receptor 2-Gi protein in complex with agonist WIN 55,212-2
Method: single particle / : Xu TH, Xing C, Zhuang Y, Feng Z, Zhou XE, Chen M, Wang L, Meng X, Xue Y, Wang J, Liu H, McGuire T, Zhao G, Melcher K, Zhang C, Xu HE, Xie XQ
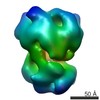
EMDB-2522: 
Substrate Recruitment Pathways in the Yeast Exosome by Electron Microscopy
Method: single particle / : Liu JJ, Bratkowski MA, Liu XQ, Niu CY, Ke AL, Wang HW

EMDB-2491: 
Recruitment Pathways in the Yeast Exosome by Electron Microscopy
Method: single particle / : Liu JJ, Bratkowski MA, Liu XQ, Niu CY, Ke AL, Wang HW

EMDB-2492: 
Substrate Recruitment Pathways in the Yeast Exosome by Electron Microscopy
Method: single particle / : Liu JJ, Bratkowski MA, Liu XQ, Niu CY, Ke AL, Wang HW

EMDB-2493: 
Substrate Recruitment Pathways in the Yeast Exosome by Electron Microscopy
Method: single particle / : Liu JJ, Bratkowski MA, Liu XQ, Niu CY, Ke AL, Wang HW

EMDB-2494: 
Substrate Recruitment Pathways in the Yeast Exosome by Electron Microscopy
Method: single particle / : Liu JJ, Bratkowski MA, Liu XQ, Niu CY, Ke AL, Wang HW

EMDB-2495: 
Substrate Recruitment Pathways in the Yeast Exosome by Electron Microscopy
Method: single particle / : Liu JJ, Bratkowski MA, Liu XQ, Niu CY, Ke AL, Wang HW

EMDB-2496: 
Substrate Recruitment Pathways in the Yeast Exosome by Electron Microscopy
Method: single particle / : Liu JJ, Bratkowski MA, Liu XQ, Niu CY, Ke AL, Wang HW

EMDB-2497: 
Substrate Recruitment Pathways in the Yeast Exosome by Electron Microscopy
Method: single particle / : Liu JJ, Bratkowski MA, Liu XQ, Niu CY, Ke AL, Wang HW

EMDB-2498: 
Substrate Recruitment Pathways in the Yeast Exosome by Electron Microscopy
Method: single particle / : Liu JJ, Bratkowski MA, Liu XQ, Niu CY, Ke AL, Wang HW
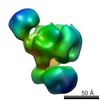
EMDB-2499: 
Substrate Recruitment Pathways in the Yeast Exosome by Electron Microscopy
Method: single particle / : Liu JJ, Bratkowski MA, Liu XQ, Niu CY, Ke AL, Wang HW
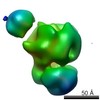
EMDB-2500: 
Substrate Recruitment Pathways in the Yeast Exosome by Electron Microscopy
Method: single particle / : Liu JJ, Bratkowski MA, Liu XQ, Niu CY, Ke AL, Wang HW
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
