-Search query
-Search result
Showing 1 - 50 of 288 items for (author: lins & b)

EMDB-18991: 
Sub-tomogram average of the C. elegans ATP synthase dimer
Method: subtomogram averaging / : Buzzard EJ, McLaren M, Gold VAM

EMDB-18329: 
yeast cytoplasmic exosome-Ski2 complex degrading a RNA substrate
Method: single particle / : Keidel A, Koegel A, Reichelt P, Kowalinski E, Schaefer IB, Conti E

PDB-8qcf: 
yeast cytoplasmic exosome-Ski2 complex degrading a RNA substrate
Method: single particle / : Keidel A, Koegel A, Reichelt P, Kowalinski E, Schaefer IB, Conti E
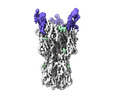
EMDB-17724: 
CryoEM reconstruction of hemagglutinin HK68 of Influenza A virus bound to an Affimer reagent
Method: single particle / : Debski-Antoniak O, Flynn A, Klebl DP, Tiede C, Muench S, Tomlinson D, Fontana J

PDB-8pk3: 
CryoEM reconstruction of hemagglutinin HK68 of Influenza A virus bound to an Affimer reagent
Method: single particle / : Debski-Antoniak O, Flynn A, Klebl DP, Tiede C, Muench S, Tomlinson D, Fontana J

EMDB-42990: 
DNA initiation complex (configuration 1) of Xenopus laevis DNA polymerase alpha-primase
Method: single particle / : Mullins EA, Durie CL, Ohi MD, Chazin WJ, Eichman BF

EMDB-42991: 
DNA initiation complex (configuration 2) of Xenopus laevis DNA polymerase alpha-primase
Method: single particle / : Mullins EA, Durie CL, Ohi MD, Chazin WJ, Eichman BF

EMDB-42992: 
DNA elongation complex (configuration 1) of Xenopus laevis DNA polymerase alpha-primase
Method: single particle / : Mullins EA, Durie CL, Ohi MD, Chazin WJ, Eichman BF

EMDB-42993: 
DNA elongation complex (configuration 2) of Xenopus laevis DNA polymerase alpha-primase
Method: single particle / : Mullins EA, Durie CL, Ohi MD, Chazin WJ, Eichman BF

PDB-8v5m: 
Tetramer core subcomplex (conformation 1) of Xenopus laevis DNA polymerase alpha-primase
Method: single particle / : Mullins EA, Chazin WJ, Eichman BF

PDB-8v5n: 
Tetramer core subcomplex (conformation 2) of Xenopus laevis DNA polymerase alpha-primase
Method: single particle / : Mullins EA, Chazin WJ, Eichman BF

PDB-8v5o: 
Tetramer core subcomplex (conformation 3) of Xenopus laevis DNA polymerase alpha-primase
Method: single particle / : Mullins EA, Chazin WJ, Eichman BF

PDB-8v6g: 
DNA initiation complex (configuration 1) of Xenopus laevis DNA polymerase alpha-primase
Method: single particle / : Mullins EA, Durie CL, Ohi MD, Chazin WJ, Eichman BF

PDB-8v6h: 
DNA initiation complex (configuration 2) of Xenopus laevis DNA polymerase alpha-primase
Method: single particle / : Mullins EA, Durie CL, Ohi MD, Chazin WJ, Eichman BF

PDB-8v6i: 
DNA elongation complex (configuration 1) of Xenopus laevis DNA polymerase alpha-primase
Method: single particle / : Mullins EA, Durie CL, Ohi MD, Chazin WJ, Eichman BF

PDB-8v6j: 
DNA elongation complex (configuration 2) of Xenopus laevis DNA polymerase alpha-primase
Method: single particle / : Mullins EA, Durie CL, Ohi MD, Chazin WJ, Eichman BF

EMDB-16942: 
Murine type II Abeta fibril from APP23 mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

EMDB-16944: 
Murine type III Abeta fibril from APP/PS1 mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

EMDB-16949: 
Murine type II Abeta fibril from ARTE10 mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

EMDB-16952: 
Murine type II Abeta fibril from tgAPPSwe mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

EMDB-16953: 
MurineArc type I Abeta fibril from tg-APPArcSwe mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

EMDB-16957: 
DI2 Abeta fibril from tg-SwDI mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

EMDB-16959: 
DI1 Abeta fibril from tg-SwDI mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

EMDB-16960: 
Murine type III Abeta fibril from ARTE10 mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

EMDB-16961: 
DI3 Abeta fibril from tg-SwDI mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

PDB-8ol2: 
Murine type II Abeta fibril from APP23 mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

PDB-8ol3: 
Murine type III Abeta fibril from APP/PS1 mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

PDB-8ol5: 
Murine type II Abeta fibril from ARTE10 mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

PDB-8ol6: 
Murine type II Abeta fibril from tgAPPSwe mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

PDB-8ol7: 
MurineArc type I Abeta fibril from tg-APPArcSwe mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

PDB-8olg: 
DI2 Abeta fibril from tg-SwDI mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

PDB-8oln: 
DI1 Abeta fibril from tg-SwDI mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

PDB-8olo: 
Murine type III Abeta fibril from ARTE10 mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

PDB-8olq: 
DI3 Abeta fibril from tg-SwDI mouse
Method: helical / : Zielinski M, Peralta Reyes FS, Gremer L, Schemmert S, Frieg B, Willuweit A, Donner L, Elvers M, Nilsson LNG, Syvanen S, Sehlin D, Ingelsson M, Willbold D, Schroeder GF

EMDB-42034: 
RNA priming complex of Human polymerase alpha-primase (Conformation 2)
Method: single particle / : Cordoba JJ, Chazin WJ

EMDB-42036: 
Human Polymerase alpha-Primase, polymerase alpha catalytic domain deletion mutant (Conformation 1)
Method: single particle / : Cordoba JJ, Chazin WJ
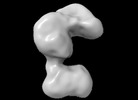
EMDB-42037: 
Human Polymerase alpha-Primase, polymerase alpha catalytic domain deletion mutant (Conformation 2)
Method: single particle / : Cordoba JJ, Chazin WJ

EMDB-18288: 
CryoEM structure of a S. Cerevisiae Ski238 complex bound to RNA
Method: single particle / : Keidel A, Koegel A, Reichelt P, Kowalinski E, Schaefer IB, Conti E

PDB-8q9t: 
CryoEM structure of a S. Cerevisiae Ski238 complex bound to RNA
Method: single particle / : Keidel A, Koegel A, Reichelt P, Kowalinski E, Schaefer IB, Conti E
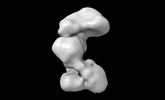
EMDB-42033: 
RNA priming complex of Human Polymerase-Alpha-Primase (Conformation 1)
Method: single particle / : Cordoba JJ, Chazin WJ
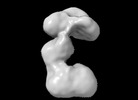
EMDB-42035: 
RNA priming complex of Human Polymerase alpha-primase (Conformation 3)
Method: single particle / : Cordoba JJ, Chazin WJ

EMDB-18326: 
CryoEM structure of a S. Cerevisiae Ski2387 complex in the closed state bound to RNA
Method: single particle / : Keidel A, Koegel A, Reichelt P, Kowalinski E, Schaefer IB, Conti E

PDB-8qca: 
CryoEM structure of a S. Cerevisiae Ski2387 complex in the closed state bound to RNA
Method: single particle / : Keidel A, Koegel A, Reichelt P, Kowalinski E, Schaefer IB, Conti E

EMDB-18327: 
CryoEM structure of a S. Cerevisiae Ski2387 complex in an intermediate state bound to RNA
Method: single particle / : Keidel A, Koegel A, Reichelt P, Kowalinski E, Schaefer IB, Conti E

EMDB-18328: 
CryoEM structure of a S. Cerevisiae Ski2387 complex in the open state
Method: single particle / : Keidel A, Koegel A, Reichelt P, Kowalinski E, Schaefer IB, Conti E

PDB-8qcb: 
CryoEM structure of a S. Cerevisiae Ski2387 complex in the open state
Method: single particle / : Keidel A, Koegel A, Reichelt P, Kowalinski E, Schaefer IB, Conti E

EMDB-42140: 
Partial DNA termination subcomplex of Xenopus laevis DNA polymerase alpha-primase
Method: single particle / : Mullins EA, Chazin WJ, Eichman BF

EMDB-42141: 
Complete DNA termination subcomplex 1 of Xenopus laevis DNA polymerase alpha-primase
Method: single particle / : Mullins EA, Chazin WC, Eichman BF

EMDB-42142: 
Complete DNA termination subcomplex 2 of Xenopus laevis DNA polymerase alpha-primase
Method: single particle / : Mullins EA, Chazin WC, Eichman BF

PDB-8ucu: 
Partial DNA termination subcomplex of Xenopus laevis DNA polymerase alpha-primase
Method: single particle / : Mullins EA, Chazin WJ, Eichman BF
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
