-Search query
-Search result
Showing 1 - 50 of 390 items for (author: liao & h)

EMDB-35769: 
ABCG25 Wild Type purified with DDM plus CHS in ABA-bound state
Method: single particle / : Sun L, Liu X, Ying W, Liao L, Wei H

PDB-8iwk: 
ABCG25 Wild Type purified with DDM plus CHS in ABA-bound state
Method: single particle / : Sun L, Liu X, Ying W, Liao L, Wei H

EMDB-35768: 
ABCG25 Wild Type in Apo-state
Method: single particle / : Sun L, Liu X, Ying W, Liao L, Wei H

PDB-8iwj: 
ABCG25 Wild Type in Apo-state
Method: single particle / : Sun L, Liu X, Ying W, Liao L, Wei H

EMDB-35774: 
ABCG25 EQ mutant in ATP-bound state
Method: single particle / : Sun L, Liu X, Ying W, Liao L, Wei H

PDB-8iwn: 
ABCG25 EQ mutant in ATP-bound state
Method: single particle / : Sun L, Liu X, Ying W, Liao L, Wei H

EMDB-37850: 
Cryo-EM structure of native H. thermoluteolus TH-1 GroEL
Method: single particle / : Liao Z, Gopalasingam CC, Kameya M, Gerle C, Shigematsu H, Ishii M, Arakawa T, Fushinobu S

EMDB-37853: 
Cryo-EM structure of H. thermoluteolus GroEL-GroES2 football complex
Method: single particle / : Liao Z, Gopalasingam CC, Kameya M, Gerle C, Shigematsu H, Ishii M, Arakawa T, Fushinobu S

EMDB-37862: 
Cryo-EM structure of H. thermophilus GroEL-GroES2 asymmetric football complex
Method: single particle / : Liao Z, Gopalasingam CC, Kameya M, Gerle C, Shigematsu H, Ishii M, Arakawa T, Fushinobu S

EMDB-37863: 
Cryo-EM structure of H. thermophilus GroEL-GroES bullet complex
Method: single particle / : Liao Z, Gopalasingam CC, Kameya M, Gerle C, Shigematsu H, Ishii M, Arakawa T, Fushinobu S

PDB-8wu4: 
Cryo-EM structure of native H. thermoluteolus TH-1 GroEL
Method: single particle / : Liao Z, Gopalasingam CC, Kameya M, Gerle C, Shigematsu H, Ishii M, Arakawa T, Fushinobu S

PDB-8wuc: 
Cryo-EM structure of H. thermoluteolus GroEL-GroES2 football complex
Method: single particle / : Liao Z, Gopalasingam CC, Kameya M, Gerle C, Shigematsu H, Ishii M, Arakawa T, Fushinobu S

PDB-8wuw: 
Cryo-EM structure of H. thermophilus GroEL-GroES2 asymmetric football complex
Method: single particle / : Liao Z, Gopalasingam CC, Kameya M, Gerle C, Shigematsu H, Ishii M, Arakawa T, Fushinobu S

PDB-8wux: 
Cryo-EM structure of H. thermophilus GroEL-GroES bullet complex
Method: single particle / : Liao Z, Gopalasingam CC, Kameya M, Gerle C, Shigematsu H, Ishii M, Arakawa T, Fushinobu S

EMDB-37849: 
Cryo-EM structure of CB1-beta-arrestin1 complex
Method: single particle / : Liao Y, Zhang H, Shen Q, Cai C

PDB-8wu1: 
Cryo-EM structure of CB1-beta-arrestin1 complex
Method: single particle / : Liao Y, Zhang H, Shen Q, Cai C

EMDB-28809: 
Yeast ATP synthase map in conformation-1 at pH 6
Method: single particle / : Sharma S, Patel H, Luo M, Mueller DM, Liao M

EMDB-28835: 
Yeast ATP synthase map in conformation-2, at pH 6
Method: single particle / : Sharma S, Patel H, Luo M, Mueller DM, Liao M

PDB-8f29: 
Yeast ATP synthase in conformation-1 at pH 6
Method: single particle / : Sharma S, Patel H, Luo M, Mueller DM, Liao M

PDB-8f39: 
Yeast ATP synthase in conformation-2, at pH 6
Method: single particle / : Sharma S, Patel H, Luo M, Mueller DM, Liao M

EMDB-29250: 
Yeast ATP Synthase in conformation-3, at pH 6
Method: single particle / : Sharma S, Patel H, Luo M, Mueller DM, Liao M

PDB-8fkj: 
Yeast ATP Synthase in conformation-3, at pH 6
Method: single particle / : Sharma S, Patel H, Luo M, Mueller DM, Liao M

EMDB-28685: 
Yeast ATP synthase consensus map in conformation-0 at pH 6
Method: single particle / : Sharma S, Patel H, Luo M, Mueller DM, Liao M

EMDB-29270: 
Yeast ATP Synthase map in presence of MgATP
Method: single particle / : Sharma S, Patel H, Luo M, Mueller DM, Liao M

EMDB-29278: 
F1-focused map of yeast ATP Synthase in presence of MgATP
Method: single particle / : Sharma S, Patel H, Luo M, Mueller DM, Liao M

PDB-8fl8: 
Yeast ATP Synthase structure in presence of MgATP
Method: single particle / : Sharma S, Patel H, Luo M, Mueller DM, Liao M

EMDB-29251: 
Fo and central rotor focused map of yeast ATP Synthase in conformation-3, at pH 6
Method: single particle / : Sharma S, Patel H, Luo M, Mueller DM, Liao M

EMDB-40223: 
Cryo-ET reconstruction of WT HSV-1 NEC coat
Method: subtomogram averaging / : Wang H, Draganova EB

EMDB-40224: 
Cryo-ET reconstruction of HSV-1 DN/SUP mutant NEC coat
Method: subtomogram averaging / : Wang H, Draganova EB

EMDB-40225: 
Cryo-ET reconstruction of HSV-1 SUP mutant NEC coat
Method: subtomogram averaging / : Wang H, Draganova EB
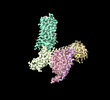
EMDB-35297: 
Structural insights into human brain gut peptide cholecystokinin receptors
Method: single particle / : Ding Y, Zhang H, Liao Y, Chen L, Ji S

PDB-8ia7: 
Structural insights into human brain gut peptide cholecystokinin receptors
Method: single particle / : Ding Y, Zhang H, Liao Y, Chen L, Ji S

EMDB-28811: 
F1-focused map of yeast ATP synthase in conformation-1, at pH 6
Method: single particle / : Sharma S, Patel H, Luo M, Mueller DM, Liao M

EMDB-28814: 
Fo and central rotor focused map of yeast ATP synthase in conformation-1, at pH 6
Method: single particle / : Sharma S, Patel H, Luo M, Mueller DM, Liao M

EMDB-28836: 
F1-focused map of yeast ATP synthase in conformation-2, at pH 6
Method: single particle / : Sharma S, Patel H, Luo M, Mueller DM, Liao M

EMDB-28837: 
Fo and central rotor focused map of yeast ATP synthase in conformation-2, at pH 6
Method: single particle / : Sharma S, Patel H, Luo M, Mueller DM, Liao M

EMDB-28813: 
Fo-focused map of yeast ATP synthase in conformation-1, at pH 6
Method: single particle / : Sharma S, Patel H, Luo M, Mueller DM, Liao M

EMDB-28687: 
Yeast ATP synthase F1-focused map in conformation-0 at pH 6
Method: single particle / : Sharma S, Patel H, Luo M, Mueller DM, Liao M

EMDB-28689: 
Yeast ATP synthase Fo-focused map in conformation-0 at pH 6
Method: single particle / : Sharma S, Patel H, Luo M, Mueller DM, Liao M

EMDB-42149: 
S1V2-72 Fab bound to EHA2 from influenza B/Malaysia/2506/2004
Method: single particle / : Finney J, Kong S, Walsh Jr RM, Harrison SC, Kelsoe G

PDB-8udg: 
S1V2-72 Fab bound to EHA2 from influenza B/Malaysia/2506/2004
Method: single particle / : Finney J, Kong S, Walsh Jr RM, Harrison SC, Kelsoe G

EMDB-41066: 
Human VMAT2 in complex with tetrabenazine
Method: single particle / : Pidathala S, Dai Y, Lee CH

EMDB-41067: 
Human VMAT2 in complex with reserpine
Method: single particle / : Pidathala S, Dai Y, Lee CH

EMDB-41068: 
Human VMAT2 in complex with serotonin
Method: single particle / : Pidathala S, Dai Y, Lee CH

PDB-8t69: 
Human VMAT2 in complex with tetrabenazine
Method: single particle / : Pidathala S, Dai Y, Lee CH

PDB-8t6a: 
Human VMAT2 in complex with reserpine
Method: single particle / : Pidathala S, Dai Y, Lee CH

PDB-8t6b: 
Human VMAT2 in complex with serotonin
Method: single particle / : Pidathala S, Dai Y, Lee CH

EMDB-29442: 
Subtomogram of the 4-beta-ring hoop of M. hungatei sheath structure.
Method: subtomogram averaging / : Wang H, Zhang J, Zhou ZH

EMDB-29443: 
Subtomogram average of 3-beta-ring hoop structure of M. hungatei sheath
Method: subtomogram averaging / : Wang H, Zhang J, Zhou ZH

EMDB-29448: 
Subtomogram average of 5-beta-ring hoop structure of M. hungatei sheath
Method: subtomogram averaging / : Wang H, Zhang J, Zhou ZH
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
