-Search query
-Search result
Showing 1 - 50 of 3,494 items for (author: lei & z)

EMDB entry, No image
EMDB-37240: 
SARS-CoV-2 Omicron spike in complex with 5817 Fab
Method: single particle / : Cao L, Wang X

EMDB entry, No image
EMDB-37241: 
The interface structure of Omicron RBD binding to 5817 Fab
Method: single particle / : Cao L, Wang X

PDB-8khc: 
SARS-CoV-2 Omicron spike in complex with 5817 Fab
Method: single particle / : Cao L, Wang X

PDB-8khd: 
The interface structure of Omicron RBD binding to 5817 Fab
Method: single particle / : Cao L, Wang X

EMDB entry, No image
EMDB-17974: 
Pseudorabies virus cytosolic C-capsid (US3 KO) vertices determined in situ
Method: subtomogram averaging / : Prazak V, Grange M, Vasishtan D

EMDB entry, No image
EMDB-17975: 
Pseudorabies virus primary enveloped (perinuclear) C-capsid (US3 KO) vertices determined in situ
Method: subtomogram averaging / : Prazak V, Grange M, Vasishtan D

EMDB entry, No image
EMDB-17976: 
Pseudorabies nuclear C-capsids (US3 KO) vertices determined in situ
Method: subtomogram averaging / : Prazak V, Grange M, Vasishtan D

EMDB entry, No image
EMDB-18473: 
Subtomogram average of pseudorabies virus nuclear egress complex helical form (UL31/34) determined in situ
Method: subtomogram averaging / : Prazak V, Grange M, Vasishtan D

EMDB entry, No image
EMDB-18474: 
Subtomogram average of pseudorabies virus nuclear egress complex (UL31/34) determined in situ
Method: subtomogram averaging / : Prazak V, Grange M, Vasishtan D

EMDB entry, No image
EMDB-18479: 
Pseudorabies virus cytosolic C-capsid (WT) vertices determined in situ
Method: subtomogram averaging / : Mironova Y, Prazak V, Vasishtan D

EMDB entry, No image
EMDB-18480: 
Pseudorabies virus nuclear C-capsid (WT) vertices determined in situ
Method: subtomogram averaging / : Mironova Y, Prazak V, Vasishtan D

EMDB entry, No image
EMDB-18481: 
Herpes simplex virus 1 cytosolic C-capsid (WT) vertices determined in situ
Method: subtomogram averaging / : Mironova Y, Prazak V, Vasishtan D

EMDB entry, No image
EMDB-18483: 
Herpes simplex virus 1 nuclear C-capsid (WT) vertices determined in situ
Method: subtomogram averaging / : Mironova Y, Prazak V, Vasishtan D

EMDB-35906: 
cryo-EM structure of human EMC
Method: single particle / : Li M, Zhang C, Wu J, Lei M
EMDB-35907: 
cryo-EM structure of human EMC and VDAC
Method: single particle / : Li M, Zhang C, Wu J, Lei M

PDB-8j0o: 
cryo-EM structure of human EMC and VDAC
Method: single particle / : Li M, Zhang C, Wu J, Lei M
EMDB-36808: 
Cryo-EM structure of KEOPS complex from Arabidopsis thaliana
Method: single particle / : Zheng XX, Zhu L, Duan L, Zhang WH

PDB-8k20: 
Cryo-EM structure of KEOPS complex from Arabidopsis thaliana
Method: single particle / : Zheng XX, Zhu L, Duan L, Zhang WH

EMDB-18110: 
The fibrillar and amorphous states of polyQ Q97
Method: electron tomography / : Zhao DY

EMDB-18114: 
phagophore in fibrillar polyQ
Method: electron tomography / : Zhao DY
EMDB-18115: 
phagophore and lysosomes with amorphous polyQ
Method: electron tomography / : Zhao DY
EMDB-43732: 
momSalB bound Kappa Opioid Receptor in complex Gi1
Method: single particle / : Fay JF, Che T

EMDB-43733: 
GR89,696 bound Kappa Opioid Receptor in complex with Gz
Method: single particle / : Fay JF, Che T

EMDB-43734: 
GR89,696 bound Kappa Opioid Receptor in complex with gustducin
Method: single particle / : Fay JF, Che T

EMDB-36678: 
PSI-AcpPCI supercomplex from Amphidinium carterae
Method: single particle / : Li ZH, Li XY, Wang WD

EMDB-36742: 
PSI-AcpPCI supercomplex from Symbiodinium
Method: single particle / : Li ZH, Li XY, Wang WD

EMDB-36743: 
PSI-AcpPCI supercomplex from Symbiodinium
Method: single particle / : Li XY, Li ZH, Wang WD

PDB-8jw0: 
PSI-AcpPCI supercomplex from Amphidinium carterae
Method: single particle / : Li ZH, Li XY, Wang WD

PDB-8jze: 
PSI-AcpPCI supercomplex from Symbiodinium
Method: single particle / : Li ZH, Li XY, Wang WD

PDB-8jzf: 
PSI-AcpPCI supercomplex from Symbiodinium
Method: single particle / : Li XY, Li ZH, Wang WD

EMDB-34314: 
SARS-CoV-2 RNA E-RTC complex with RMP-nsp9 and GMPPNP
Method: single particle / : Yan LM, Huang YC, Ge J, Liu ZY, Gao Y, Rao ZH, Lou ZY

EMDB-34316: 
SARS-CoV-2 E-RTC bound with MMP-nsp9 and GMPPNP
Method: single particle / : Yan LM, Rao ZH, Lou ZY

PDB-8gwk: 
SARS-CoV-2 RNA E-RTC complex with RMP-nsp9 and GMPPNP
Method: single particle / : Yan LM, Huang YC, Ge J, Liu ZY, Gao Y, Rao ZH, Lou ZY

PDB-8gwm: 
SARS-CoV-2 E-RTC bound with MMP-nsp9 and GMPPNP
Method: single particle / : Yan LM, Rao ZH, Lou ZY

EMDB-18181: 
Early closed conformation of the g-tubulin ring complex
Method: single particle / : Llorca O, Serna M, Fernandez-Leiro R

PDB-8q62: 
Early closed conformation of the g-tubulin ring complex
Method: single particle / : Llorca O, Serna M, Fernandez-Leiro R

EMDB-16781: 
NTD focused cryo-EM map of p97/VCP in ADP.Pi state
Method: single particle / : Cheng TC, Sakata E, Schuetz AK

EMDB-17016: 
Full composite cryo-EM map of p97/VCP in ADP.Pi state
Method: single particle / : Cheng TC, Sakata E, Schuetz AK

EMDB-17024: 
D1-D2 ring focused cryo-EM map of p97/VCP in ADP.Pi state
Method: single particle / : Cheng TC, Sakata E, Schuetz AK

EMDB-17128: 
Consensus cryo-EM map of p97/VCP in ADP.Pi state
Method: single particle / : Cheng TC, Sakata E, Schuetz AK

PDB-8ooi: 
Full composite cryo-EM map of p97/VCP in ADP.Pi state
Method: single particle / : Cheng TC, Sakata E, Schuetz AK

EMDB-42602: 
Campylobacter jejuni CosR apo form
Method: single particle / : Zhang Z
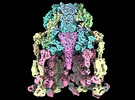
EMDB-29383: 
Structure of baseplate with receptor binding complex of Agrobacterium phage Milano
Method: single particle / : Sonani RR, Leiman PG, Wang F, Kreutzberger MAB, Sebastian A, Esteves NC, Kelly RJ, Scharf B, Egelman EH

PDB-8fqc: 
Structure of baseplate with receptor binding complex of Agrobacterium phage Milano
Method: single particle / : Sonani RR, Leiman PG, Wang F, Kreutzberger MAB, Sebastian A, Esteves NC, Kelly RJ, Scharf B, Egelman EH

EMDB-42804: 
Structure of nucleotide-free Pediculus humanus (Ph) PINK1 dimer
Method: single particle / : Gan ZY, Kirk NS, Leis A, Komander D

EMDB-42806: 
Structure of AMP-PNP-bound Pediculus humanus (Ph) PINK1 dimer
Method: single particle / : Gan ZY, Kirk NS, Leis A, Komander D

EMDB-42807: 
Structure of ADP-bound and phosphorylated Pediculus humanus (Ph) PINK1 dimer
Method: single particle / : Gan ZY, Kirk NS, Leis A, Komander D

PDB-8uyf: 
Structure of nucleotide-free Pediculus humanus (Ph) PINK1 dimer
Method: single particle / : Gan ZY, Kirk NS, Leis A, Komander D
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
