-Search query
-Search result
Showing 1 - 50 of 65 items for (author: lee & lj)

EMDB-28850: 
SARS-CoV-2 Gamma 6P Mut7 S + COVA309-3 Fab
Method: single particle / : Torres JL, Ward AB

EMDB-28851: 
SARS-CoV-2 Gamma 6P Mut7 S + COVA309-10 Fab
Method: single particle / : Torres JL, Ward AB

EMDB-28852: 
SARS-CoV-2 Omicron 6P S + COVA309-35 Fab
Method: single particle / : Torres JL, Ward AB
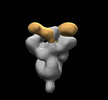
EMDB-28853: 
SARS-CoV-2 Gamma 6P Mut7 + S COVA309-38 Fab
Method: single particle / : Torres JL, Ward AB

EMDB-28755: 
Human tRNA Splicing Endonuclease Complex bound to 2'F-tRNA-Arg
Method: single particle / : Stanley RE, Hayne CK
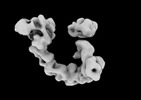
EMDB-29101: 
EGFR:Degrader:VHL:Elongin-B/C:Cul2
Method: single particle / : Rosenberg SC

EMDB-26856: 
Human tRNA Splicing Endonuclease Complex bound to pre-tRNA-ARG
Method: single particle / : Stanley RE, Hayne CK

EMDB-26831: 
Human Rix1 sub-complex scaffold
Method: single particle / : Gordon J, Stanley RE
EMDB-13586: 
Cryo-electron tomography of ASC signalling sites in pyroptotic cells (2)
Method: electron tomography / : Liu Y, Modis Y, Alemayehu H, Hopkins LJ, Borgeaud AC, Heroven C, Howe JD, Boulanger J, Bryant CE, Zhai H

EMDB-25474: 
CryoEM structure of the N-terminal-deleted Rix7 AAA-ATPase
Method: single particle / : Kocaman S, Stanley RE, Lo YH, Krahn J, Dandey VP, Sobhany M, Petrovich M, Williams JG, Deterding LJ, Borgnia MJ, Etigunta S

EMDB-25582: 
CryoEM structure of the crosslinked Rix7 AAA-ATPase
Method: single particle / : Kocaman S, Stanley RE, Lo YH, Krahn J, Dandey VP, Sobhany M, Petrovich M, Williams JG, Deterding LJ, Borgnia MJ, Etigunta S

EMDB-25659: 
CryoEM structure of the Rix7 D2 Walker B mutant
Method: single particle / : Lo YH, Krahn J

EMDB-23708: 
ATP-bound AMP-activated protein kinase
Method: single particle / : Yan Y, Mukherjee S, Harikumar KG, Strutzenberg T, Zhou XE, Powell SK, Xu T, Sheldon R, Lamp J, Brunzelle JS, Radziwon K, Ellis A, Novick SJ, Vega IE, Jones R, Miller LJ, Xu HE, Griffin PR, Kossiakoff AA, Melcher K

EMDB-24642: 
SARS-CoV-2 Spike bound to Fab PDI 210
Method: single particle / : Pymm P, Glukhova A, Black K, Tham WH

EMDB-24643: 
SARS-CoV-2 Spike bound to Fab PDI 96
Method: single particle / : Pymm P, Glukhova A, Black K, Tham WH

EMDB-24644: 
SARS-CoV-2 Spike bound to Fab PDI 215
Method: single particle / : Black K, Glukhova A, Pymm P, Tham WH

EMDB-24645: 
SARS-CoV-2 Spike bound to Fab WCSL 119
Method: single particle / : Black K, Glukhova A, Pymm P, Tham WH

EMDB-24646: 
SARS-CoV-2 Spike bound to Fab WCSL 129
Method: single particle / : Black K, Glukhova A, Pymm P, Tham WH

EMDB-24647: 
SARS-CoV-2 Spike bound to Fab PDI 93
Method: single particle / : Black K, Glukhova A, Pymm P, Tham WH

EMDB-24648: 
SARS-CoV-2 Spike bound to Fab PDI 222
Method: single particle / : Glukhova A, Pymm P, Black K, Tham WH

EMDB-24649: 
SARS-CoV-2 receptor binding domain bound to Fab PDI 222
Method: single particle / : Pymm P, Glukhova A, Black KA, Tham WH

EMDB-22336: 
Cryo-EM structure of ATP-bound fully inactive AMPK in complex with Dorsomorphin (Compound C) and Fab-nanobody
Method: single particle / : Yan Y, Murkherjee S, Zhou XE, Xu TH, Xu HE, Kossiakoff AA, Melcher K

EMDB-22337: 
Cryo-EM structure of ATP-bound fully inactive AMPK in complex with Fab and nanobody
Method: single particle / : Yan Y, Murkherjee S, Zhou XE, Xu TH, Xu HE, Kossiakoff AA, Melcher K

EMDB-24137: 
SARS-CoV-2 Nsp15 endoribonuclease pre-cleavage state
Method: single particle / : Frazier MN, Dillard LB, Krahn JM, Stanley RE

EMDB-24101: 
SARS-CoV-2 Nsp15 endoribonuclease post-cleavage state
Method: single particle / : Frazier MN, Dillard LB, Krahn JM, Stanley RE

EMDB-23749: 
Human Cholecystokinin 1 receptor (CCK1R) Gs complex
Method: single particle / : Mobbs JI, Belousoff MJ, Danev R, Thal DM, Sexton PM

EMDB-23750: 
Human Cholecystokinin 1 receptor (CCK1R) Gq chimera (mGsqi) complex
Method: single particle / : Mobbs JI, Belousoff MJ, Danev R, Thal DM, Sexton PM

EMDB-12286: 
Cryo-EM structure of the ternary complex between Netrin-1, Neogenin and Repulsive Guidance Molecule B
Method: single particle / : Robinson RA, Griffiths SC, van de Haar LL, Malinauskas T, van Battum EY, Zelina P, Schwab RA, Karia D, Malinauskaite L, Brignani S, van den Munkhof M, Dudukcu O, De Ruiter AA, Van den Heuvel DMA, Bishop B, Elegheert J, Aricescu AR, Pasterkamp RJ, Siebold C

PDB-7ndg: 
Cryo-EM structure of the ternary complex between Netrin-1, Neogenin and Repulsive Guidance Molecule B
Method: single particle / : Robinson RA, Griffiths SC, van de Haar LL, Malinauskas T, van Battum EY, Zelina P, Schwab RA, Karia D, Malinauskaite L, Brignani S, van den Munkhof M, Dudukcu O, De Ruiter AA, Van den Heuvel DMA, Bishop B, Elegheert J, Aricescu AR, Pasterkamp RJ, Siebold C

EMDB-23042: 
Chimeric flavivirus between Binjari virus and Dengue virus serotype-2
Method: single particle / : Hardy JM, Venugopal HV, Newton ND, Watterson D, Coulibaly FJ

EMDB-23043: 
Chimeric flavivirus between Binjari virus and West Nile (Kunjin) virus
Method: single particle / : Hardy JM, Venugopal HV, Newton ND, Watterson D, Coulibaly FJ

EMDB-23044: 
Structure of West Nile virus (Kunjin)
Method: single particle / : Hardy JM, Venugopal HV, Newton ND, Watterson D, Coulibaly FJ

EMDB-23045: 
Chimeric flavivirus between Binjari virus and Murray Valley encephalitis virus
Method: single particle / : Hardy JM, Venugopal HV, Newton ND, Watterson D, Coulibaly FJ

EMDB-22610: 
Nucleotide bound SARS-CoV-2 Nsp15
Method: single particle / : Pillon MC, Stanley RE

EMDB-22611: 
SARS-CoV-2 wt-Nsp15 APO-state
Method: single particle / : Pillon MC, Stanley RE

EMDB-22612: 
SARS-CoV-2 Nsp15 H235A variant APO-state, dataset ii
Method: single particle / : Pillon MC, Stanley RE

EMDB-22613: 
SARS-CoV-2 Nsp15 H235A APO-state, dataset i
Method: single particle / : Pillon MC, Stanley RE

EMDB-22829: 
Human Tom70 in complex with SARS CoV2 Orf9b
Method: single particle / : QCRG Structural Biology Consortium

PDB-7kdt: 
Human Tom70 in complex with SARS CoV2 Orf9b
Method: single particle / : QCRG Structural Biology Consortium

EMDB-21683: 
Human secretin receptor Gs complex
Method: single particle / : Belousoff MJ, Khoshouei M

EMDB-21972: 
Human secretin receptor Gs complex
Method: single particle / : Piper SJ, Belousoff MJ, Danev R

EMDB-20417: 
Cryo-EM reconstruction of the mature chimeric BinJV/ZIKV-prME virion
Method: single particle / : Watterson D

EMDB-20438: 
Cryo-EM reconstruction of the mature chimeric BinJV/ZIKV-prME virion in complex with Fab C8
Method: single particle / : Watterson D

EMDB-20439: 
Cryo-EM reconstruction of the immature chimeric BinJV/ZIKV-prME virion
Method: single particle / : Watterson D

EMDB-9757: 
Whole structure of a 15-stranded ParM filament from Clostridium botulinum
Method: helical / : Koh F, Narita A, Lee LJ, Tan YZ, Dandey VP, Tanaka K, Popp D, Robinson RC

EMDB-9758: 
Averaged strand structure of a 15-stranded ParM filament from Clostridium botulinum
Method: helical / : Koh F, Narita A, Lee LJ, Tan YZ, Dandey VP, Tanaka K, Popp D, Robinson RC

EMDB-0350: 
Induction of Potent Neutralizing Antibody Responses by a Designed Protein Nanoparticle Vaccine for Respiratory Syncytial Virus
Method: single particle / : Marcandalli J, Fiala B, Ols S, Perotti M, van der Schueren W, Snijder S, Hodge E, Benhaim M, Ravichandran R, Carter L, Sheffler W, Brunner L, Lawrenz M, Dubois P, Lanzavecchia A, Sallusto F, Lee KK, Veesler D, Correnti CE, Stewart LJ, Baker D, Lore K, Perez L, King NP

EMDB-7092: 
Cryo-EM structure of human insulin degrading enzyme in complex with insulin
Method: single particle / : Zhang Z, Liang WG, Bailey LJ, Tan YZ, Wei H, Kossiakoff AA, Carragher B, Potter SC, Tang WJ

EMDB-7090: 
Cryo-EM structure of human insulin degrading enzyme
Method: single particle / : Zhang Z, Liang WG, Bailey LJ, Tan YZ, Wei H, Kossiakoff AA, Carragher B, Potter SC, Tang WJ

EMDB-7091: 
Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain
Method: single particle / : Zhang Z, Liang WG, Bailey LJ, Tan YZ, Wei H, Kossiakoff AA, Carragher B, Potter SC, Tang WJ
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
