-Search query
-Search result
Showing all 33 items for (author: lau & wc)

EMDB-37631: 
Hepatitis B virus capsid (HBV core protein)
Method: single particle / : Yip RPH, Lai LTF, Lau WCY, Ngo JCK, Kwok DCY

EMDB-37634: 
SR protein kinase 2 bound at 2-fold vertex of Hepatitis B virus capsid
Method: single particle / : Yip RPH, Lai LTF, Kwok DCY, Lau WCY, Ngo JCK

EMDB-38062: 
Hepatitis B virus capsid in complex with SR protein kinase 2
Method: single particle / : Yip RPH, Lai LTF, Kwok DCY, Lau WCY, Ngo JCK

EMDB-42141: 
Complete DNA termination subcomplex 1 of Xenopus laevis DNA polymerase alpha-primase
Method: single particle / : Mullins EA, Chazin WC, Eichman BF

EMDB-42142: 
Complete DNA termination subcomplex 2 of Xenopus laevis DNA polymerase alpha-primase
Method: single particle / : Mullins EA, Chazin WC, Eichman BF

EMDB-26667: 
Cryo-EM structure of SHOC2-PP1c-MRAS holophosphatase complex
Method: single particle / : Fuller JR, Hajian B, Lemke C, Kwon J, Bian Y, Aguirre A

EMDB-25263: 
SARS-CoV-2 S B.1.617.2 delta variant + S2M11 + S2L20 Global Refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-25264: 
SARS-CoV-2 S NTD B.1.617.2 delta variant + S2L20 Local Refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-25265: 
SARS-CoV-2 S B.1.617.1 kappa variant + S309 + S2L20 Global Refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-25266: 
SARS-CoV-2 S RBD B.1.617.1 kappa variant S309 Local Refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-25267: 
SARS-CoV-2 S NTD B.1.617.1 kappa variant S2L20 Local Refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-25268: 
SARS-CoV-2 S B.1.617.1 kappa variant + S2X303 Global Refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-25269: 
SARS-CoV-2 S NTD B.1.617.1 kappa variant S2X303 Local Refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-30262: 
DXPS
Method: single particle / : Lau WCY

EMDB-30263: 
Hsp21-DXPS
Method: single particle / : Lau WCY

EMDB-24236: 
SARS-CoV-2 S (B.1.429 / epsilon variant) + S2M11 + S2L20 Global Refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-24237: 
SARS-CoV-2 S (B.1.429 / epsilon variant) + S2M11 + S2L20 (Local Refinement of the NTD/S2L20)
Method: single particle / : McCallum M, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-30261: 
Structure of Hsp21
Method: single particle / : Lau WCY

EMDB-9681: 
Arabidopsis thaliana ATG9
Method: single particle / : Lai LTF, Yu C, Wong JSK, Lo HS, Benlekbir S, Jiang L, Lau WCY

EMDB-6499: 
Structural insights into the regulatory mechanism of ATM kinase activation
Method: single particle / : Lau WCY, Li Y, Liu Z, Gao Y, Zhang Q, Huen MSY
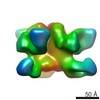
EMDB-6500: 
Negative stain EM reconstruction of HIV-1 Env AMC008 SOSIP.v4 in complex with Fabs 35O22 and PGV04
Method: single particle / : de Taeye SW, Ozorowski G, de la Pena AT, Guttman M, Julien JP, van den Kerkhof TLGM, Burger J, Pritchard LK, Pugach P, Yasmeen A, Bontjer I, Torres JL, Arendt H, DeStefano J, Koff WC, Schuitemaker H, Eggink D, Berkhout B, Dean H, LaBranche C, Crispin M, Montefiori DC, Klasse PJ, Lee KK, Moore JP, Wilson IA, Ward AB, Sanders RW

EMDB-6226: 
Substrate-free TRiC
Method: single particle / : Roh SH, Kasembeli M, Montoya JG, Trnka M, Lau WC, Burlingame A, Chiu W, Tweardy DJ

EMDB-6227: 
AML1-175-bound TRiC
Method: single particle / : Roh SH, Kasembeli M, Montoya JG, Trnka M, Lau WC, Burlingame A, Chiu W, Tweardy DJ

EMDB-6228: 
Human apo-TRiC
Method: single particle / : Roh SH, Kasembeli M, Montoya JG, Trnka M, Lau WC, Burlingame A, Chiu W, Tweardy DJ

EMDB-6229: 
Human apo-TRiC (sub-class II)
Method: single particle / : Roh SH, Kasembeli M, Montoya JG, Trnka M, Lau WC, Burlingame A, Chiu W, Tweardy DJ

EMDB-6230: 
AML1-175 + TRiC + Ni-nano-gold
Method: single particle / : Roh SH, Kasembeli M, Montoya JG, Trnka M, Lau WC, Burlingame A, Chiu W, Tweardy DJ

EMDB-6339: 
Molecular architecture of the Ub-PCNA/Pol eta complex bound to DNA
Method: single particle / : Lau WCY, Li Y, Zhang Q, Huen MSY

EMDB-2011: 
Structure of the mitochondrial ATP synthase from Saccharomyces cerevisiae in Brij-35 at 24 Angstroms resolution.
Method: single particle / : Lau WCY, Baker LA, Rubinstein JL

EMDB-5335: 
Sub-nanometer resolution structure of the intact T. thermophilus proton-driven ATP synthase reveals the arrangement of its trans-membrane helices
Method: single particle / : Lau WCY, Rubinstein JL

EMDB-1970: 
Negative stained image reconstruction of HIV spike protein in complex with PGT128 Fab at 14 Angstrom resolution
Method: single particle / : Khayat R, Pejchal R, Julien JP, Wilson IA
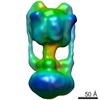
EMDB-1888: 
Thermus thermophilus V-type (A-type) ATPase at 16 Angstroms resolution
Method: single particle / : Lau WCY, Rubinstein JL
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
