-Search query
-Search result
Showing 1 - 50 of 143 items for (author: lara & s)
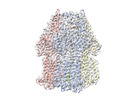
EMDB-17350: 
Single particle cryo-EM co-structure of Klebsiella pneumoniae AcrB with the BDM91288 efflux pump inhibitor at 2.97 Angstrom resolution
Method: single particle / : Boernsen C, Mueller RT, Pos KM, Frangakis AS

PDB-8p1i: 
Single particle cryo-EM co-structure of Klebsiella pneumoniae AcrB with the BDM91288 efflux pump inhibitor at 2.97 Angstrom resolution
Method: single particle / : Boernsen C, Mueller RT, Pos KM, Frangakis AS
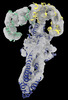
EMDB-18149: 
GBP1 bound by 14-3-3sigma
Method: single particle / : Pfleiderer MM, Liu X, Fisch D, Anastasakou E, Frickel EM, Galej WP

PDB-8q4l: 
GBP1 bound by 14-3-3sigma
Method: single particle / : Pfleiderer MM, Liu X, Fisch D, Anastasakou E, Frickel EM, Galej WP
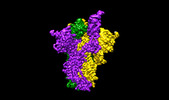
EMDB-17576: 
SARS-CoV-2 S-protein:D614G mutant in 1-up conformation
Method: single particle / : Adhav A, Forcada-Nadal A, Marco-Marin C, Lopez-Redondo ML, Llacer JL
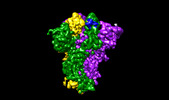
EMDB-17578: 
SARS-CoV-2 S protein S:D614G mutant in 3-down with binding site of an entry inhibitor
Method: single particle / : Adhav A, Forcada-Nadal A, Marco-Marin C, Lopez-Redondo ML, Llacer JL

PDB-8p99: 
SARS-CoV-2 S-protein:D614G mutant in 1-up conformation
Method: single particle / : Adhav A, Forcada-Nadal A, Marco-Marin C, Lopez-Redondo ML, Llacer JL

PDB-8p9y: 
SARS-CoV-2 S protein S:D614G mutant in 3-down with binding site of an entry inhibitor
Method: single particle / : Adhav A, Forcada-Nadal A, Marco-Marin C, Lopez-Redondo ML, Llacer JL

EMDB-41624: 
Cryo-EM structure of the human MRS2 magnesium channel under Mg2+ condition
Method: single particle / : Lai LTF, Balaraman J, Zhou F, Matthies D

EMDB-41628: 
Cryo-EM structure of the human MRS2 magnesium channel under Mg2+-free condition
Method: single particle / : Lai LTF, Balaraman J, Zhou F, Matthies D

EMDB-41629: 
Cryo-EM structure of the human MRS2 magnesium channel under Mg2+ condition (C1 map)
Method: single particle / : Lai LTF, Balaraman J, Zhou F, Matthies D

EMDB-41630: 
Cryo-EM structure of the human MRS2 magnesium channel under Mg2+-free condition (C1 map)
Method: single particle / : Lai LTF, Balaraman J, Zhou F, Matthies D

PDB-8tul: 
Cryo-EM structure of the human MRS2 magnesium channel under Mg2+ condition
Method: single particle / : Lai LTF, Balaraman J, Zhou F, Matthies D

PDB-8tup: 
Cryo-EM structure of the human MRS2 magnesium channel under Mg2+-free condition
Method: single particle / : Lai LTF, Balaraman J, Zhou F, Matthies D

EMDB-15403: 
Structure of the secreted Yersinia entomophaga Tc toxin YenTc
Method: subtomogram averaging / : Sitsel O, Wang Z, Janning P, Kroczek L, Raunser S

EMDB-15404: 
Tomogram of Yersinia entomophaga in the pre-secretory state
Method: electron tomography / : Sitsel O, Wang Z, Janning P, Kroczek L, Raunser S

EMDB-15405: 
Tomogram of Yersinia entomophaga in the post-holin state
Method: electron tomography / : Sitsel O, Wang Z, Janning P, Kroczek L, Raunser S
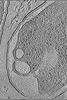
EMDB-15406: 
Tomogram of Yersinia entomophaga in the post-endolysin state
Method: electron tomography / : Sitsel O, Wang Z, Janning P, Kroczek L, Raunser S
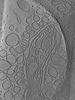
EMDB-15407: 
Tomogram of Yersinia entomophaga in the post-spanin state
Method: electron tomography / : Sitsel O, Wang Z, Janning P, Kroczek L, Raunser S

EMDB-41046: 
Subtomogram average structure of the injectisome
Method: subtomogram averaging / : Park D
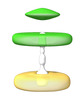
EMDB-41047: 
Intermembrane Tether between SCV and Mitochondria
Method: subtomogram averaging / : Park D, Liu J, Galan J

EMDB-15700: 
Type 3 secretion system needle complex of Shigella flexneri
Method: single particle / : Lunelli M

EMDB-15701: 
Outer membrane secretin pore of the type 3 secretion system of Shigella flexneri
Method: single particle / : Lunelli M

EMDB-15702: 
Inner membrane ring and secretin N0 N1 domains of the type 3 secretion system of Shigella flexneri
Method: single particle / : Lunelli M

EMDB-15703: 
Inner membrane ring of the type 3 secretion system of Shigella flexneri
Method: single particle / : Lunelli M

PDB-8axk: 
Type 3 secretion system export apparatus core, inner rod and needle of Shigella flexneri
Method: single particle / : Lunelli M

PDB-8axl: 
Outer membrane secretin pore of the type 3 secretion system of Shigella flexneri
Method: single particle / : Lunelli M

PDB-8axn: 
Inner membrane ring and secretin N0 N1 domains of the type 3 secretion system of Shigella flexneri
Method: single particle / : Lunelli M
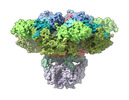
EMDB-27982: 
Structure of the full-length IP3R1 channel determined at high Ca2+
Method: single particle / : Fan G, Baker MR, Terry LE, Arige V, Chen M, Seryshev AB, Baker ML, Ludtke SJ, Yule DI, Serysheva II
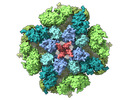
EMDB-27983: 
Structure of the full-length IP3R1 channel determined in the presence of Calcium/IP3/ATP
Method: single particle / : Fan G, Baker MR, Terry LE, Arige V, Chen M, Seryshev AB, Baker ML, Ludtke SJ, Yule DI, Serysheva II

PDB-8eaq: 
Structure of the full-length IP3R1 channel determined at high Ca2+
Method: single particle / : Fan G, Baker MR, Terry LE, Arige V, Chen M, Seryshev AB, Baker ML, Ludtke SJ, Yule DI, Serysheva II

PDB-8ear: 
Structure of the full-length IP3R1 channel determined in the presence of Calcium/IP3/ATP
Method: single particle / : Fan G, Baker MR, Terry LE, Arige V, Chen M, Seryshev AB, Baker ML, Ludtke SJ, Yule DI, Serysheva II

EMDB-26604: 
H1 Solomon Islands 2006 hemagglutinin in complex with Ab111
Method: single particle / : Windsor IW, Caradonna TM, Schmidt AG
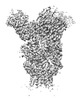
EMDB-26605: 
H1 Solomon Islands 2006 hemagglutinin in complex with Ab109
Method: single particle / : Windsor IW, Caradonna TM, Schmidt AG

PDB-7umm: 
H1 Solomon Islands 2006 hemagglutinin in complex with Ab109
Method: single particle / : Windsor IW, Caradonna TM, Schmidt AG

EMDB-13546: 
Sulfolobus acidocaldarius 0406 filament.
Method: helical / : Isupov MN, Gaines M, Daum B

EMDB-14472: 
Structure of the RAF1-HSP90-CDC37 complex (RHC-II)
Method: single particle / : Mesa P, Garcia-Alonso S, Barbacid M, Montoya G

EMDB-14473: 
Structure of the RAF1-HSP90-CDC37 complex (RHC-I)
Method: single particle / : Mesa P, Garcia-Alonso S, Barbacid M, Montoya G

PDB-7z37: 
Structure of the RAF1-HSP90-CDC37 complex (RHC-II)
Method: single particle / : Mesa P, Garcia-Alonso S, Barbacid M, Montoya G

PDB-7z38: 
Structure of the RAF1-HSP90-CDC37 complex (RHC-I)
Method: single particle / : Mesa P, Garcia-Alonso S, Barbacid M, Montoya G

EMDB-25039: 
Structure of H1 NC99 influenza hemagglutinin bound to Fab 310-63E6
Method: single particle / : Ward A, Torrents de la Pena A

EMDB-25040: 
Structure of H1 influenza hemagglutinin bound to Fab 310-39G10
Method: single particle / : Torrents de la Pena A, Ward AB

PDB-7scn: 
Structure of H1 NC99 influenza hemagglutinin bound to Fab 310-63E6
Method: single particle / : Ward A, Torrents de la Pena A

PDB-7sco: 
Structure of H1 influenza hemagglutinin bound to Fab 310-39G10
Method: single particle / : Torrents de la Pena A, Ward AB
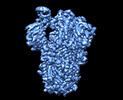
EMDB-13916: 
SARS-CoV-2 S protein S:A222V + S:D614G mutant 1-up
Method: single particle / : Ginex T, Marco-Marin C, Wieczor M, Mata CP, Krieger J, Lopez-Redondo ML, Frances-Gomez C, Ruiz-Rodriguez P, Melero R, Sanchez-Sorzano CO, Martinez M, Gougeard N, Forcada-Nadal A, Zamora-Caballero S, Gozalbo-Rovira R, Sanz-Frasquet C, Bravo J, Rubio V, Marina A, Geller R, Comas I, Gil C, Coscolla M, Orozco M, LLacer JL, Carazo JM
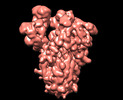
EMDB-13917: 
SARS-CoV-2 S protein S:A222V + S:D614G mutant 2-up
Method: single particle / : Ginex T, Marco-Marin C, Wieczor M, Mata CP, Krieger J, Lopez-Redondo ML, Ruiz-Rodriguez P, Melero R, Sanchez-Sorzano CO, Martinez M, Gougeard N, Forcada-Nadal A, Zamora-Caballero S, Gozalbo-Rovira R, Sanz-Frasquet C, Bravo J, Rubio V, Marina A, Geller R, Comas I, Gil C, Coscolla M, Orozco M, Llacer JL, Carazo JM
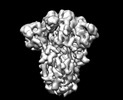
EMDB-13918: 
SARS-CoV-2 S protein S:A222V + S:D614G mutant 3-down
Method: single particle / : Ginex T, Marco-Marin C, Wieczor M, Mata CP, Krieger J, Lopez-Redondo ML, Ruiz-Rodriguez P, Melero R, Sanchez-Sorzano CO, Martinez M, Gougeard N, Forcada-Nadal A, Zamora-Caballero S, Gozalbo-Rovira R, Sanz-Frasquet C, Bravo J, Rubio V, Marina A, Geller R, Comas I, Gil C, Coscolla M, Orozco M, Llacer JL, Carazo JM
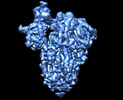
EMDB-13919: 
SARS-CoV-2 S protein S:D614G mutant 1-up
Method: single particle / : Ginex T, Marco-Marin C, Wieczor M, Mata CP, Krieger J, Lopez-Redondo ML, Frances-Gomez C, Ruiz-Rodriguez P, Melero R, Sanchez-Sorzano CO, Martinez M, Gougeard N, Forcada-Nadal A, Zamora-Caballero S, Gozalbo-Rovira R, Sanz-Frasquet C, Bravo J, Rubio V, Marina A, Geller R, Comas I, Gil C, Coscolla M, Orozco M, LLacer JL, Carazo JM
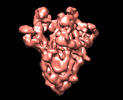
EMDB-13920: 
SARS-CoV-2 S protein S:D614G mutant 2-up
Method: single particle / : Ginex T, Marco-Marin C, Wieczor M, Mata CP, Krieger J, Lopez-Redondo ML, Ruiz-Rodriguez P, Melero R, Sanchez-Sorzano CO, Martinez M, Gougeard N, Forcada-Nadal A, Zamora-Caballero S, Gozalbo-Rovira R, Sanz-Frasquet C, Bravo J, Rubio V, Marina A, Geller R, Comas I, Gil C, Coscolla M, Orozco M, Llacer JL, Carazo JM
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
