-Search query
-Search result
Showing 1 - 50 of 894 items for (author: kumar & m)

EMDB entry, No image
EMDB-41105: 
CryoEM structure of human DDB1-DCAF12 in complex with MAGEA3
Method: single particle / : Duda D, Righetto G, Li Y, Loppnau P, Seitova A, Santhakumar V, Halabelian L, Yin Y

PDB-8t9a: 
CryoEM structure of human DDB1-DCAF12 in complex with MAGEA3
Method: single particle / : Duda D, Righetto G, Li Y, Loppnau P, Seitova A, Santhakumar V, Halabelian L, Yin Y

EMDB-18942: 
Structure of coxsackievirus B5 capsid (mutant CVB5F.cas.genogroupB) - F particle
Method: single particle / : Kumar K, Antanasijevic A

EMDB-18943: 
Structure of coxsackievirus B5 capsid (mutant CVB5F.cas.genogroupB) - A particle
Method: single particle / : Kumar K, Antanasijevic A

EMDB-18944: 
Structure of coxsackievirus B5 capsid (mutant CVB5F.cas.genogroupB) - E particle
Method: single particle / : Kumar K, Antanasijevic A

PDB-8r5x: 
Structure of coxsackievirus B5 capsid (mutant CVB5F.cas.genogroupB) - F particle
Method: single particle / : Kumar K, Antanasijevic A

PDB-8r5y: 
Structure of coxsackievirus B5 capsid (mutant CVB5F.cas.genogroupB) - A particle
Method: single particle / : Kumar K, Antanasijevic A

PDB-8r5z: 
Structure of coxsackievirus B5 capsid (mutant CVB5F.cas.genogroupB) - E particle
Method: single particle / : Kumar K, Antanasijevic A

EMDB-36331: 
Atomic structure of wheat ribosome reveals unique features of the plant ribosomes
Method: single particle / : Mishra RK, Sharma P, Hussain T

EMDB-36332: 
Atomic structure of wheat ribosome reveals unique features of the plant ribosomes
Method: single particle / : Mishra RK, Sharma P, Hussain T

EMDB-36333: 
Atomic structure of wheat ribosome reveals unique features of the plant ribosomes
Method: single particle / : Mishra RK, Sharma PN, Hussain T

EMDB-36334: 
Atomic structure of wheat ribosome reveals unique features of the plant ribosomes
Method: single particle / : Mishra RK, Sharma PN, Hussain T

PDB-8jiv: 
Atomic structure of wheat ribosome reveals unique features of the plant ribosomes
Method: single particle / : Mishra RK, Sharma P, Hussain T

PDB-8jiw: 
Atomic structure of wheat ribosome reveals unique features of the plant ribosomes
Method: single particle / : Mishra RK, Sharma P, Hussain T

EMDB-43091: 
hGBP1 conformer on the bacterial outer membrane
Method: subtomogram averaging / : Zhu S, MacMicking J

EMDB-43153: 
hGBP1 conformer on the bacterial outer membrane
Method: subtomogram averaging / : Zhu S, MacMicking J

EMDB-42601: 
CryoEM structure of Kappa Opioid Receptor bound to a semi-peptide and Gi1
Method: single particle / : Fay JF, Che T

EMDB-29950: 
SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 002-S21B10
Method: single particle / : Patel A, Ortlund EA

EMDB-29975: 
Overall map of SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 002-S21B10
Method: single particle / : Patel A, Ortlund EA

EMDB-40007: 
Local map of SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 002-S21B10
Method: single particle / : Patel A, Ortlund EA

EMDB-37551: 
Cryo- EM structure of Mycobacterium smegmatis 70S ribosome and RafH.
Method: single particle / : Kumar N, Sharma S, Kaushal PS

EMDB-37552: 
Cryo- EM structure of Mycobacterium smegmatis 50S ribosomal subunit (body 1) of 70S ribosome and RafH.
Method: single particle / : Kumar N, Sharma S, Kaushal PS

EMDB-37559: 
Cryo- EM structure of Mycobacterium smegmatis 70S ribosome, bS1 and RafH.
Method: single particle / : Kumar N, Sharma S, Kaushal PS

EMDB-37560: 
Cryo- EM structure of Mycobacterium smegmatis 50S ribosomal subunit (body 1) of 70S ribosome, bS1 and RafH.
Method: single particle / : Kumar N, Sharma S, Kaushal PS

EMDB-37561: 
Cryo- EM structure of Mycobacterium smegmatis 30S ribosomal subunit (body 2) of 70S ribosome, bS1 and RafH.
Method: single particle / : Kumar N, Sharma S, Kaushal PS

EMDB-37562: 
Cryo- EM structure of Mycobacterium smegmatis 70S ribosome, E- tRNA and RafH.
Method: single particle / : Kumar N, Sharma S, Kaushal PS

EMDB-37563: 
Cryo- EM structure of Mycobacterium smegmatis 50S ribosomal subunit (body 1) of 70S ribosome, E- tRNA and RafH.
Method: single particle / : Kumar N, Sharma S, Kaushal PS

EMDB-37564: 
Cryo- EM structure of Mycobacterium smegmatis 30S ribosomal subunit (body 2) of 70S ribosome, E- tRNA and RafH.
Method: single particle / : Kumar N, Sharma S, Kaushal PS

EMDB-37565: 
Cryo- EM structure of Mycobacterium smegmatis 30S ribosomal subunit (body 2) of 70S ribosome and RafH.
Method: single particle / : Kumar N, Sharma S, Kaushal PS

PDB-8whx: 
Cryo- EM structure of Mycobacterium smegmatis 70S ribosome and RafH.
Method: single particle / : Kumar N, Sharma S, Kaushal PS

PDB-8why: 
Cryo- EM structure of Mycobacterium smegmatis 50S ribosomal subunit (body 1) of 70S ribosome and RafH.
Method: single particle / : Kumar N, Sharma S, Kaushal PS

PDB-8wi7: 
Cryo- EM structure of Mycobacterium smegmatis 70S ribosome, bS1 and RafH.
Method: single particle / : Kumar N, Sharma S, Kaushal PS

PDB-8wi8: 
Cryo- EM structure of Mycobacterium smegmatis 50S ribosomal subunit (body 1) of 70S ribosome, bS1 and RafH.
Method: single particle / : Kumar N, Sharma S, Kaushal PS

PDB-8wi9: 
Cryo- EM structure of Mycobacterium smegmatis 30S ribosomal subunit (body 2) of 70S ribosome, bS1 and RafH.
Method: single particle / : Kumar N, Sharma S, Kaushal PS

PDB-8wib: 
Cryo- EM structure of Mycobacterium smegmatis 70S ribosome, E- tRNA and RafH.
Method: single particle / : Kumar N, Sharma S, Kaushal PS

PDB-8wic: 
Cryo- EM structure of Mycobacterium smegmatis 50S ribosomal subunit (body 1) of 70S ribosome, E- tRNA and RafH.
Method: single particle / : Kumar N, Sharma S, Kaushal PS

PDB-8wid: 
Cryo- EM structure of Mycobacterium smegmatis 30S ribosomal subunit (body 2) of 70S ribosome, E- tRNA and RafH.
Method: single particle / : Kumar N, Sharma S, Kaushal PS

PDB-8wif: 
Cryo- EM structure of Mycobacterium smegmatis 30S ribosomal subunit (body 2) of 70S ribosome and RafH.
Method: single particle / : Kumar N, Sharma S, Kaushal PS
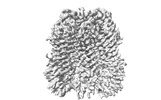
EMDB-34266: 
CryoEM structure of human Pannexin1 with R217H congenital mutation.
Method: single particle / : Hussain N, Penmatsa A

EMDB-34267: 
Cryo-EM structure of human Pannexin1 resembling Pannexin2 pore with W74R/R75Dmutations
Method: single particle / : Hussain N, Penmatsa A

EMDB-29898: 
Cannabinoid receptor 1-Gi complex with novel ligand
Method: single particle / : Tummino TA, Iliopoulos-Tsoutsouvas C, Braz JM, O'Brien ES, Krishna Kumar K, Makriyannis M, Basbaum AI, Shoichet BK

PDB-8gag: 
Cannabinoid receptor 1-Gi complex with novel ligand
Method: single particle / : Tummino TA, Iliopoulos-Tsoutsouvas C, Braz JM, O'Brien ES, Krishna Kumar K, Makriyannis M, Basbaum AI, Shoichet BK

EMDB-34265: 
CryoEM structure of human Pannexin isoform 3
Method: single particle / : Hussain N, Penmatsa A
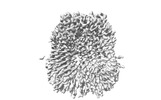
EMDB-34268: 
human Pannexin1
Method: single particle / : Hussain N, Penmatsa A, Vinothkumar KR

EMDB-36619: 
Structure of the 30S-IF3 complex from Escherichia coli
Method: single particle / : Uday AB, Mishra RK, Hussain T

PDB-8jsg: 
Structure of the 30S-IF3 complex from Escherichia coli
Method: single particle / : Uday AB, Mishra RK, Hussain T

EMDB-42185: 
Cryo-EM structure of POmAb, a Type-I anti-prothrombin antiphospholipid antibody, bound to kringle-1 of human prothrombin
Method: single particle / : Kumar S, Summers B, Basore K, Pozzi N

PDB-8uf7: 
Cryo-EM structure of POmAb, a Type-I anti-prothrombin antiphospholipid antibody, bound to kringle-1 of human prothrombin
Method: single particle / : Kumar S, Summers B, Basore K, Pozzi N

EMDB-35304: 
Cryo-EM structure of the yeast SPT-ORM2 (ORM2-S3A) complex
Method: single particle / : Xie T, Gong X

EMDB-35306: 
Cryo-EM structure of the yeast SPT-ORM2 (ORM2-S3A-N71A) complex
Method: single particle / : Xie T, Gong X
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
