-Search query
-Search result
Showing all 50 items for (author: kjaer & s)

EMDB-29911: 
The Type 9 Secretion System Extended Translocon - delta GldL, Peak I, SprA-PorV-RemZ-PPI focused volume
Method: single particle / : Deme JC, Lea SM
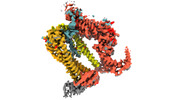
EMDB-40085: 
The Type 9 Secretion System Extended Translocon - delta GldL, Peak I, SprE-SkpA-Nterm SprA focused volume
Method: single particle / : Deme JC, Lea SM

EMDB-40086: 
The Type 9 Secretion System Extended Translocon - delta GldL, Peak I, consensus volume
Method: single particle / : Deme JC, Lea SM

EMDB-40191: 
The Type 9 Secretion System in vitro assembled, RemA-CTD substrate bound complex
Method: single particle / : Deme JC, Lea SM

EMDB-40194: 
The Type 9 Secretion System Extended Translocon - SprA-PorV-PPI-RemZ-SkpA-SprE complex
Method: single particle / : Deme JC, Lea SM

EMDB-40195: 
The Type 9 Secretion System in vitro assembled, FspA-CTD substrate bound complex
Method: single particle / : Deme JC, Lea SM

EMDB-40196: 
The Type 9 Secretion System dGldL peak II, NucA substrate bound complex
Method: single particle / : Deme JC, Lea SM

EMDB-40199: 
The Type 9 Secretion System in vivo assembled, RemZ substrate bound complex - conformation 1
Method: single particle / : Deme JC, Lea SM

EMDB-40201: 
The Type 9 Secretion System in vivo assembled, RemZ substrate bound complex - conformation 2
Method: single particle / : Deme JC, Lea SM

EMDB-15217: 
PAPP-A dimer in complex with a dimer of the inhibitor STC2
Method: single particle / : Kobbero SD, Oxvig C, Gajhede M, Boesen T

EMDB-15219: 
PAPP-A dimer in complex with endogenous STC2 inhibitor.
Method: single particle / : Kobbero SD, Oxvig C, Gajhede M, Boesen T

EMDB-15220: 
Partial dimer complex of PAPP-A and its inhibitor STC2
Method: single particle / : Kobbero SD, Gajhede M, Mirza OA, Boesen T, Oxvig C

EMDB-15221: 
PAPP-A dimer in complex with its inhibitor STC2
Method: single particle / : Kobbero SD, Gajhede M, Mirza OA, Boesen T, Oxvig C

EMDB-13355: 
Structure of the Caulobacter crescentus S-layer protein RsaA N-terminal domain bound to LPS and soaked with Holmium
Method: single particle / : von Kugelgen A, Bharat TAM

PDB-7peo: 
Structure of the Caulobacter crescentus S-layer protein RsaA N-terminal domain bound to LPS and soaked with Holmium
Method: single particle / : von Kugelgen A, Bharat TAM

EMDB-12585: 
Trimeric SARS-CoV-2 spike ectodomain in complex with biliverdin (closed conformation)
Method: single particle / : Rosa A, Pye VE, Nans A, Cherepanov P

EMDB-12586: 
Trimeric SARS-CoV-2 spike ectodomain in complex with biliverdin (one RBD erect)
Method: single particle / : Rosa A, Pye VE, Nans A, Cherepanov P

EMDB-12587: 
Trimeric SARS-CoV-2 spike ectodomain bound to P008_056 Fab
Method: single particle / : Rosa A, Pye VE, Nans A, Cherepanov P

EMDB-11777: 
RET/GDF15/GFRAL extracellular complex negative stain envelope
Method: single particle / : Adams SE, Earl CP, McDonald NQ

EMDB-11822: 
RET/GDNF/GFRa1 extracellular complex Cryo-EM structure
Method: single particle / : Adams SE, Earl CP, Purkiss AG, McDonald NQ

EMDB-10893: 
Structure of GldLM, the proton-powered motor that drives protein transport and gliding motility
Method: single particle / : Hennell James R, Deme JC, Lea SM

EMDB-10959: 
Cryo-EM structure of the ARP2/3 1B5CL isoform complex.
Method: single particle / : von Loeffelholz O, Moores C, Purkiss A

EMDB-10960: 
Cryo-EM structure of the ARP2/3 1A5C isoform complex.
Method: single particle / : von Loeffelholz O, Moores C, Purkiss A

PDB-6yw6: 
Cryo-EM structure of the ARP2/3 1B5CL isoform complex.
Method: single particle / : von Loeffelholz O, Moores C, Purkiss A

PDB-6yw7: 
Cryo-EM structure of the ARP2/3 1A5C isoform complex.
Method: single particle / : von Loeffelholz O, Moores C, Purkiss A

EMDB-4605: 
Human Phenylalanine Hydroxylase (hPAH) apo structure
Method: single particle / : Alcorlo-Pages M, Flydal MI, Martinez-Caballero S, Martinez A, Hermoso JA, Fernandez-Leiro R

EMDB-0464: 
Legionella pneumophila bacterial flagellar motor
Method: subtomogram averaging / : Kaplan M, Ghosal D, Subramanian P, Oikonomou CO, Kjaer A, Pirbadian S, Ortega DR, Briegel A, El-Naggar MY, Jensen GJ

EMDB-0465: 
Pseudomonas aeruginosa bacterial flagellar motor
Method: subtomogram averaging / : Kaplan M, Ghosal D, Subramanian P, Oikonomou CO, Kjaer A, Pirbadian S, Ortega DR, Briegel A, El-Naggar MY, Jensen GJ
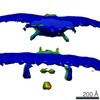
EMDB-0467: 
Shewanella oneidensis MR-1 bacterial flagellar motor
Method: subtomogram averaging / : Kaplan M, Ghosal D, Subramanian P, Oikonomou CO, Kjaer A, Pirbadian S, Ortega DR, Briegel A, El-Naggar MY, Jensen GJ

EMDB-3792: 
Electron cryotomogram of Vibrio cholerae O395 N1
Method: electron tomography / : Chang YW, Kjaer A, Ortega DR, Kovacikova G, Sutherland JA, Rettberg LA, Taylor RK, Jensen GJ

EMDB-8603: 
Sub-tomogram average of the archaellar motor in Thermococcus kodakaraensis
Method: subtomogram averaging / : Chang YW, Kjaer A, Briegel A, Oikonomou CM, Jensen GJ

EMDB-8492: 
Subtomogram average of the non-piliated toxin-coregulated pilus machine in wild-type Vibrio cholerae cells (aligning OM-parts)
Method: subtomogram averaging / : Chang YW, Kjaer A, Jensen GJ

EMDB-8493: 
Subtomogram average of the non-piliated toxin-coregulated pilus machine in wild-type Vibrio cholerae cells (aligning IM-parts)
Method: subtomogram averaging / : Chang YW, Kjaer A, Jensen GJ

EMDB-8494: 
Subtomogram average of the piliated toxin-coregulated pilus machine in wild-type Vibrio cholerae cells (aligning OM-parts)
Method: subtomogram averaging / : Chang YW, Kjaer A, Jensen GJ

EMDB-8495: 
Subtomogram average of the piliated toxin-coregulated pilus machine in wild-type Vibrio cholerae cells (aligning IM-parts)
Method: subtomogram averaging / : Chang YW, Kjaer A, Jensen GJ

EMDB-8496: 
Subtomogram average of the non-piliated toxin-coregulated pilus machine in Vibrio cholerae cells with tcpS knockout (aligning OM-parts)
Method: subtomogram averaging / : Chang YW, Kjaer A, Jensen GJ

EMDB-8497: 
Subtomogram average of the non-piliated toxin-coregulated pilus machine in Vibrio cholerae cells with tcpS knockout (aligning IM-parts)
Method: subtomogram averaging / : Chang YW, Kjaer A, Jensen GJ

EMDB-8498: 
Subtomogram average of the non-piliated toxin-coregulated pilus machine in Vibrio cholerae cells with tcpB knockout (aligning OM-parts)
Method: subtomogram averaging / : Chang YW, Kjaer A, Jensen GJ

EMDB-8499: 
Subtomogram average of the non-piliated toxin-coregulated pilus machine in Vibrio cholerae cells with tcpB knockout (aligning IM-parts)
Method: subtomogram averaging / : Chang YW, Kjaer A, Jensen GJ

EMDB-8500: 
Subtomogram average of the non-piliated toxin-coregulated pilus machine in Vibrio cholerae cells with tcpD knockout (aligning OM-parts)
Method: subtomogram averaging / : Chang YW, Kjaer A, Jensen GJ

EMDB-8501: 
Subtomogram average of the non-piliated toxin-coregulated pilus machine in Vibrio cholerae cells with tcpD knockout (aligning IM-parts)
Method: subtomogram averaging / : Chang YW, Kjaer A, Jensen GJ

EMDB-8502: 
Subtomogram average of the non-piliated toxin-coregulated pilus machine in Vibrio cholerae cells with tcpR knockout (aligning OM-parts)
Method: subtomogram averaging / : Chang YW, Kjaer A, Jensen GJ

EMDB-8503: 
Subtomogram average of the non-piliated toxin-coregulated pilus machine in Vibrio cholerae cells with tcpR knockout (aligning IM-parts)
Method: subtomogram averaging / : Chang YW, Kjaer A, Jensen GJ

EMDB-3398: 
Electron Cryotomography and subvolume averaging of the cytoplasmic chemoreceptor array in Vibrio cholerae
Method: subtomogram averaging / : Briegel A, Ortega D R, Mann P, Kjaer A, Ringgaard S, Jensen G J

EMDB-6102: 
Electron cryo-microscopy of DNGR-1 in complex with F-actin
Method: single particle / : Hanc P, Fujii T, Yamada Y, Huotari J, Schulz O, Ahrens S, Kjaer S, Way M, Namba K, Reis e Sousa C

PDB-3j82: 
Electron cryo-microscopy of DNGR-1 in complex with F-actin
Method: single particle / : Hanc P, Fujii T, Yamada Y, Huotari J, Schulz O, Ahrens S, Kjaer S, Way M, Namba K, Reis e Sousa C

EMDB-2712: 
Structure of the RET receptor tyrosine kinase extracellular domain
Method: single particle / : Goodman K, Kjaer S, Beuron F, Knowles P, Nawrotek A, Burns E, Purkiss A, George R, Santoro M, Morris EP, McDonald NQ

EMDB-2713: 
Structure of the zebrafish RET tyrosine kinase extracellular domain
Method: single particle / : Goodman K, Kjaer S, Beuron F, Knowles P, Nawrotek A, Burns E, Purkiss A, George R, Santoro M, Morris EP, McDonald NQ
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
