-Search query
-Search result
Showing 1 - 50 of 1,057 items for (author: kim & la)

EMDB-41888: 
Structure of Apo CXCR4/Gi complex
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC

EMDB-41889: 
Structure of CXCL12-bound CXCR4/Gi complex
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC

EMDB-41890: 
Structure of AMD3100-bound CXCR4/Gi complex
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC

EMDB-41891: 
Structure of REGN7663 Fab-bound CXCR4/Gi complex
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC

EMDB-41892: 
Structure of REGN7663-Fab bound CXCR4
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC

EMDB-41893: 
Structure of trimeric CXCR4 in complex with REGN7663 Fab
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC

EMDB-41894: 
Structure of tetrameric CXCR4 in complex with REGN7663 Fab
Method: single particle / : Saotome K, McGoldrick LL, Franklin MC
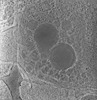
EMDB-28138: 
3D reconstruction of the apical complex of Plasmodium falciparum (3D7) free merozoite
Method: electron tomography / : Segev-Zarko L, Sun SY, Kim CY
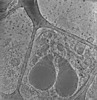
EMDB-28141: 
3D reconstruction of the apical complex of Plasmodium falciparum (3D7) free merozoite
Method: electron tomography / : Segev-Zarko L, Sun SY, Kim CY
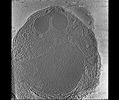
EMDB-28142: 
3D Reconstruction of Plasmodium falciparum (3D7) free merozoite
Method: electron tomography / : Segev-Zarko L, Sun SY, Kim CY

EMDB-16590: 
Structure of the yeast delta uL16m mitochondrial ribosome - State 2
Method: single particle / : Conrad J, Rathore S, Barrientos A
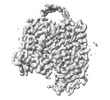
EMDB-35377: 
Cryo-EM structure of GPR156 of GPR156-miniGo-scFv16 complex (local refine)
Method: single particle / : Shin J, Park J, Cho Y

EMDB-35378: 
Cryo-EM structure of miniGo-scFv16 of GPR156-miniGo-scFv16 complex (local refine)
Method: single particle / : Shin J, Park J, Cho Y

EMDB-35380: 
Cryo-EM structure of GPR156-miniGo-scFv16 complex
Method: single particle / : Shin J, Park J, Cho Y

EMDB-35382: 
Cryo-EM structure of GPR156A/B of G-protein free GPR156 (local refine)
Method: single particle / : Shin J, Park J, Cho Y

EMDB-35389: 
Cryo-EM structure of GPR156C/D of G-protein free GPR156 (local refine)
Method: single particle / : Shin J, Park J, Cho Y

EMDB-35390: 
Cryo-EM structure of G-protein free GPR156
Method: single particle / : Shin J, Park J, Cho Y

PDB-8ieb: 
Cryo-EM structure of GPR156 of GPR156-miniGo-scFv16 complex (local refine)
Method: single particle / : Shin J, Park J, Cho Y

PDB-8iec: 
Cryo-EM structure of miniGo-scFv16 of GPR156-miniGo-scFv16 complex (local refine)
Method: single particle / : Shin J, Park J, Cho Y

PDB-8ied: 
Cryo-EM structure of GPR156-miniGo-scFv16 complex
Method: single particle / : Shin J, Park J, Cho Y

PDB-8iei: 
Cryo-EM structure of GPR156A/B of G-protein free GPR156 (local refine)
Method: single particle / : Shin J, Park J, Cho Y

PDB-8iep: 
Cryo-EM structure of GPR156C/D of G-protein free GPR156 (local refine)
Method: single particle / : Shin J, Park J, Cho Y

PDB-8ieq: 
Cryo-EM structure of G-protein free GPR156
Method: single particle / : Shin J, Park J, Cho Y

EMDB-16559: 
Structure of the yeast delta mtg1 mitochondrial ribosome assembly intermediate - State1
Method: single particle / : Conrad J, Rathore S, Barrientos A

EMDB-16560: 
Structure of the yeast delta mtg1 mitochondrial ribosome assembly intermediate - State 2
Method: single particle / : Conrad J, Rathore S, Barrientos A

EMDB-16561: 
Structure of the yeast delta mtg1 mitochondrial ribosome - State 3
Method: single particle / : Conrad J, Rathore S, Barrientos A

EMDB-16562: 
Structure of the yeast delta uL16m mitochondrial ribosome assembly intermediate - State 1
Method: single particle / : Conrad J, Rathore S, Barrientos A

EMDB-43049: 
Cryo-electron tomogram of mitochondria from cryo-FIB milled l-Opa1* mouse embryonic fibroblasts
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43050: 
cryo-electron tomogram of mitochondria in cryo-FIB milled l-Opa1* mouse embryonic fibroblasts
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Luce BE, Lugo CM, Chao LH

EMDB-43051: 
cryo-electron tomogram of mitochondria in cryo-FIB milled l-Opa1* mouse embryonic fibroblasts
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Luce BE, Lugo CM, Chao LH

EMDB-43052: 
cryo-electron tomogram of mitochondria in cryo-FIB milled l-Opa1* mouse embryonic fibroblasts
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Luce BE, Lugo CM, Chao LH

EMDB-43053: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled s-Opa1* mouse embryonic fibroblasts
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43054: 
cryo-electron tomogram of mitochondria in cryo-FIB milled s-Opa1* mouse embryonic fibroblasts
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43055: 
cryo-electron tomogram of mitochondria in cryo-FIB milled s-Opa1* mouse embryonic fibroblasts
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43056: 
cryo-electron tomogram of mitochondria in cryo-FIB milled s-Opa1* mouse embryonic fibroblasts
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43057: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled s-Opa1* mouse embryonic fibroblasts
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43058: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts with OPA1 deletion
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43059: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts with OPA1 deletion
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Luce BE, Lugo CM, Chao LH

EMDB-43060: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts with OPA1 deletion
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Luce BE, Lugo CM, Chao LH

EMDB-43061: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts with OPA1 deletion
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Luce BE, Lugo CM, Chao LH

EMDB-43062: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts with OPA1 deletion
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43063: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts
Method: electron tomography / : Navarro PP, Fry MY, Ananda VY, Ge Y, McDonald JL, Ali I, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43064: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts
Method: electron tomography / : Navarro PP, Fry MY, Ananda VY, Ge Y, McDonald JL, Ali I, Hakim P, Lugo CM, Luce BE, Chao LH
EMDB-43065: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts
Method: electron tomography / : Navarro PP, Fry MY, Ananda VY, Ge Y, McDonald JL, Ali I, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43066: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts
Method: electron tomography / : Navarro PP, Fry MY, Ananda VY, Ge Y, McDonald JL, Ali I, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43067: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts with overexpression of OPA1
Method: electron tomography / : Navarro PP, Fry MY, Ananda VY, Ge Y, McDonald JL, Ali I, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43068: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts with overexpression of OPA1
Method: electron tomography / : Navarro PP, Fry MY, Ananda VY, Ge Y, McDonald JL, Ali I, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43069: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts with overexpression of OPA1
Method: electron tomography / : Navarro PP, Fry MY, Ananda VY, Ge Y, McDonald JL, Ali I, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43070: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts with overexpression of OPA1
Method: electron tomography / : Navarro PP, Fry MY, Ananda VY, Ge Y, McDonald JL, Ali I, Hakim P, Lugo CM, Luce BE, Chao LH
EMDB-28752: 
Cryo-electron tomography of wild type a-synuclein preformed fibrils
Method: electron tomography / : Jiang J, Boparai N, Dai W, Kim YS
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
