-Search query
-Search result
Showing 1 - 50 of 162 items for (author: kay & rr)

EMDB-19276: 
Cryo-EM of tetrameric collagenase-cleaved Xenopus ZP2-N2N3 (cleaved xZP2-N2N3) (C1)
Method: single particle / : Wiseman B, Nishio S, Jovine L

EMDB-19277: 
Cryo-EM of tetrameric collagenase-cleaved Xenopus ZP2-N2N3 (cleaved xZP2-N2N3) (C2)
Method: single particle / : Wiseman B, Nishio S, Jovine L
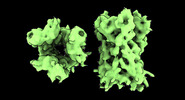
EMDB-17704: 
Subtomogram average of Vaccinia A10 trimer with open center from in vitro cores
Method: subtomogram averaging / : Turonova B, Liu J
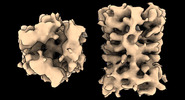
EMDB-17708: 
Subtomogram average of Vaccinia A10 trimer with tight center from in vitro cores
Method: subtomogram averaging / : Turonova B, Liu J

EMDB-17753: 
Subtomogram average of Vaccinia A10 trimer from in situ cores
Method: subtomogram averaging / : Turonova B, Liu J

EMDB-41140: 
APC/C-CDH1-UBE2C-Ubiquitin-CyclinB-NTD
Method: single particle / : Bodrug T, Welsh KA, Bolhuis DL, Paulakonis E, Martinez-Chacin RC, Liu B, Pinkin N, Bonacci T, Cui L, Xu P, Roscow O, Amann SJ, Grishkovskaya I, Emanuele MJ, Harrison JS, Steimel JP, Hahn KM, Zhang W, Zhong E, Haselbach D, Brown NG

EMDB-41142: 
APC/C-CDH1-UBE2C-UBE2S-Ubiquitin-CyclinB
Method: single particle / : Bodrug T, Welsh KA, Bolhuis DL, Paulakonis E, Martinez-Chacin RC, Liu B, Pinkin N, Bonacci T, Cui L, Xu P, Roscow O, Amann SJ, Grishkovskaya I, Emanuele MJ, Harrison JS, Steimel JP, Hahn KM, Zhang W, Zhong E, Haselbach D, Brown NG

PDB-8tar: 
APC/C-CDH1-UBE2C-Ubiquitin-CyclinB-NTD
Method: single particle / : Bodrug T, Welsh KA, Bolhuis DL, Paulakonis E, Martinez-Chacin RC, Liu B, Pinkin N, Bonacci T, Cui L, Xu P, Roscow O, Amann SJ, Grishkovskaya I, Emanuele MJ, Harrison JS, Steimel JP, Hahn KM, Zhang W, Zhong E, Haselbach D, Brown NG

PDB-8tau: 
APC/C-CDH1-UBE2C-UBE2S-Ubiquitin-CyclinB
Method: single particle / : Bodrug T, Welsh KA, Bolhuis DL, Paulakonis E, Martinez-Chacin RC, Liu B, Pinkin N, Bonacci T, Cui L, Xu P, Roscow O, Amann SJ, Grishkovskaya I, Emanuele MJ, Harrison JS, Steimel JP, Hahn KM, Zhang W, Zhong E, Haselbach D, Brown NG

EMDB-15532: 
Plasmodium falciparum sporozoite subpellicular microtubule with interrupted luminal helix determined in situ
Method: subtomogram averaging / : Prazak V, Ferreira JL, Vasishtan D, Grunewald K, Siggel M, Hentzschel F, Pietsch E, Kosinski J, Frischknecht F, Gilberger TW

EMDB-15534: 
Plasmodium falciparum gametocyte subpellicular microtubule with 13 protofilaments determined in situ
Method: subtomogram averaging / : Ferreira JL, Prazak V, Vasishtan D, Grunewald K, Siggel M, Hentzschel F, Pietsch E, Kosinski J, Frischknecht F, Gilberger TW

EMDB-15535: 
Plasmodium falciparum gametocyte subpellicular microtubule with 14 protofilaments determined in situ
Method: subtomogram averaging / : Ferreira JL, Prazak V, Vasishtan D, Grunewald K, Siggel M, Hentzschel F, Pietsch E, Kosinski J, Frischknecht F, Gilberger TW

EMDB-15536: 
Plasmodium falciparum gametocyte subpellicular microtubule with 15 protofilaments determined in situ
Method: subtomogram averaging / : Ferreira JL, Prazak V, Vasishtan D, Grunewald K, Siggel M, Hentzschel F, Pietsch E, Kosinski J, Frischknecht F, Gilberger TW

EMDB-15537: 
Plasmodium falciparum gametocyte subpellicular microtubule with 16 protofilaments determined in situ
Method: subtomogram averaging / : Ferreira JL, Prazak V, Vasishtan D, Grunewald K, Siggel M, Hentzschel F, Pietsch E, Kosinski J, Frischknecht F, Gilberger TW

EMDB-15538: 
Plasmodium falciparum gametocyte subpellicular microtubule with 17 protofilaments determined in situ
Method: subtomogram averaging / : Ferreira JL, Prazak V, Vasishtan D, Grunewald K, Siggel M, Hentzschel F, Pietsch E, Kosinski J, Frischknecht F, Gilberger TW

EMDB-15539: 
Plasmodium falciparum gametocyte subpellicular microtubule with 18 protofilaments determined in situ
Method: subtomogram averaging / : Ferreira JL, Prazak V, Vasishtan D, Grunewald K, Siggel M, Hentzschel F, Pietsch E, Kosinski J, Frischknecht F, Gilberger TW

EMDB-15607: 
Structure of the SFTSV L protein bound to 5' cRNA hook [5' HOOK]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-15608: 
Structure of the SFTSV L protein stalled at early elongation [EARLY-ELONGATION]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-15610: 
Structure of the SFTSV L protein stalled at early elongation with the endonuclease domain in a raised conformation [EARLY-ELONGATION-ENDO]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-15614: 
Structure of the SFTSV L protein stalled at late elongation [LATE-ELONGATION]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-15615: 
Structure of the SFTSV L protein bound in a resting state [RESTING]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

PDB-8as6: 
Structure of the SFTSV L protein bound to 5' cRNA hook [5' HOOK]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

PDB-8as7: 
Structure of the SFTSV L protein stalled at early elongation [EARLY-ELONGATION]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

PDB-8asb: 
Structure of the SFTSV L protein stalled at early elongation with the endonuclease domain in a raised conformation [EARLY-ELONGATION-ENDO]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

PDB-8asd: 
Structure of the SFTSV L protein stalled at late elongation [LATE-ELONGATION]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

PDB-8asg: 
Structure of the SFTSV L protein bound in a resting state [RESTING]
Method: single particle / : Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M

EMDB-29002: 
Nucleocapsid monomer structure from SARS-CoV-2
Method: single particle / : Casasanta M, Jonaid GM, Kaylor L, Luqiu W, DiCecco L, Solares M, Berry S, Kelly DF

EMDB-29072: 
SARS-CoV-2 Nucleocapsid dimer structure determined from COVID-19 patients
Method: single particle / : Casasanta M, Jonaid GM, Kaylor L, Luqiu W, DiCecco L, Solares M, Berry S, Kelly DF

PDB-8fd5: 
Nucleocapsid monomer structure from SARS-CoV-2
Method: single particle / : Casasanta M, Jonaid GM, Kaylor L, Luqiu W, DiCecco L, Solares M, Berry S, Kelly DF

PDB-8fg2: 
SARS-CoV-2 Nucleocapsid dimer structure determined from COVID-19 patients
Method: single particle / : Casasanta M, Jonaid GM, Kaylor L, Luqiu W, DiCecco L, Solares M, Berry S, Kelly DF

EMDB-33010: 
Cryo-EM structure of human subnucleosome (closed form)
Method: single particle / : Nozawa K, Takizawa Y, Kurumizaka H

EMDB-33011: 
Cryo-EM structure of human subnucleosome (open form)
Method: single particle / : Nozawa K, Takizawa Y, Kurumizaka H

EMDB-33991: 
Cryo-EM structure of human subnucleosome (intermediate form)
Method: single particle / : Nozawa K, Takizawa Y, Kurumizaka H

PDB-7x57: 
Cryo-EM structure of human subnucleosome (closed form)
Method: single particle / : Nozawa K, Takizawa Y, Kurumizaka H

PDB-7x58: 
Cryo-EM structure of human subnucleosome (open form)
Method: single particle / : Nozawa K, Takizawa Y, Kurumizaka H

PDB-7yoz: 
Cryo-EM structure of human subnucleosome (intermediate form)
Method: single particle / : Nozawa K, Takizawa Y, Kurumizaka H

EMDB-14437: 
Structure of substrate bound DRG1 (AFG2)
Method: single particle / : Prattes M, Grishkovskaya I, Bergler H, Haselbach D

EMDB-14471: 
Structure of pre-60S particle bound to DRG1(AFG2)
Method: single particle / : Prattes M, Grishkovskaya I, Bergler H, Haselbach D

PDB-7z11: 
Structure of substrate bound DRG1 (AFG2)
Method: single particle / : Prattes M, Grishkovskaya I, Bergler H, Haselbach D

PDB-7z34: 
Structure of pre-60S particle bound to DRG1(AFG2).
Method: single particle / : Prattes M, Grishkovskaya I, Bergler H, Haselbach D

EMDB-23634: 
AAV3 in liquid phase
Method: single particle / : Jonaid GM, Kelly DF

EMDB-25476: 
G32A4 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction)
Method: single particle / : Windsor IW, Tong P, Wesemann DR, Harrison SC

EMDB-25477: 
C98C7 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction)
Method: single particle / : Windsor IW, Tong P, Wesemann DR, Harrison SC

EMDB-25478: 
G32Q4 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction)
Method: single particle / : Windsor IW, Tong P, Wesemann DR, Harrison SC

PDB-7swn: 
G32A4 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction)
Method: single particle / : Windsor IW, Tong P, Wesemann DR, Harrison SC

PDB-7swo: 
C98C7 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction)
Method: single particle / : Windsor IW, Tong P, Wesemann DR, Harrison SC

PDB-7swp: 
G32Q4 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction)
Method: single particle / : Windsor IW, Tong P, Wesemann DR, Harrison SC

EMDB-12566: 
Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with HIP-B
Method: single particle / : Baronina A, Pike ACW, Yu X, Dong YY, Shintre CA, Tessitore A, Chu A, Rotty B, Venkaya S, Mukhopadhyay SMM, Borkowska O, Chalk R, Shrestha L, Burgess-Brown NA, Edwards AM, Arrowsmith CH, Bountra C, Han S, Carpenter EP, Structural Genomics Consortium (SGC)

PDB-7nsg: 
Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with HIP-B
Method: single particle / : Baronina A, Pike ACW, Yu X, Dong YY, Shintre CA, Tessitore A, Chu A, Rotty B, Venkaya S, Mukhopadhyay SMM, Borkowska O, Chalk R, Shrestha L, Burgess-Brown NA, Edwards AM, Arrowsmith CH, Bountra C, Han S, Carpenter EP, Structural Genomics Consortium (SGC)

EMDB-13416: 
Human voltage-gated potassium channel Kv3.1 (apo condition)
Method: single particle / : Chi G, Venkaya S, Singh NK, McKinley G, Mukhopadhyay SMM, Marsden B, MacLean EM, Fernandez-Cid A, Pike ACW, Savva C, Ragan TJ, Burgess-Brown NA, Duerr KL
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
