-Search query
-Search result
Showing 1 - 50 of 56 items for (author: kaji & h)

EMDB-37480: 
PSI-LHCI of the red alga Cyanidium caldarium RK-1 (NIES-2137)
Method: single particle / : Kato K, Hamaguchi T, Nakajima Y, Kawakami K, Yonekura K, Shen JR, Nagao R

PDB-8wey: 
PSI-LHCI of the red alga Cyanidium caldarium RK-1 (NIES-2137)
Method: single particle / : Kato K, Hamaguchi T, Nakajima Y, Kawakami K, Yonekura K, Shen JR, Nagao R

EMDB-34981: 
Cryo-EM structure of human NTCP-myr-preS1-YN9048Fab complex
Method: single particle / : Asami J, Shimizu T, Ohto U

EMDB-34982: 
Cryo-EM structure of human NTCP-myr-preS1-YN9016Fab complex
Method: single particle / : Asami J, Shimizu T, Ohto U

PDB-8hrx: 
Cryo-EM structure of human NTCP-myr-preS1-YN9048Fab complex
Method: single particle / : Asami J, Shimizu T, Ohto U

PDB-8hry: 
Cryo-EM structure of human NTCP-myr-preS1-YN9016Fab complex
Method: single particle / : Asami J, Shimizu T, Ohto U
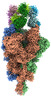
EMDB-33972: 
Cryo-EM structure of the SARS-CoV-2 spike protein (3-up RBD) bound to neutralizing antibody CSW1-1805
Method: single particle / : Anzai I, Fujita J, Ono C, Kosaka Y, Miyamoto Y, Shichinohe S, Takada K, Torii S, Taguwa S, Suzuki K, Makino F, Kajita T, Inoue T, Namba K, Watanabe T, Matsuura Y
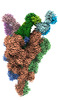
EMDB-33973: 
Cryo-EM structure of the SARS-CoV-2 spike protein (1-up RBD) bound to neutralizing antibody CSW2-1353
Method: single particle / : Anzai I, Fujita J, Ono C, Kosaka Y, Miyamoto Y, Shichinohe S, Takada K, Torii S, Taguwa S, Suzuki K, Makino F, Kajita T, Inoue T, Namba K, Watanabe T, Matsuura Y
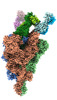
EMDB-33974: 
Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing antibody CSW2-1353
Method: single particle / : Anzai I, Fujita J, Ono C, Kosaka Y, Miyamoto Y, Shichinohe S, Takada K, Torii S, Taguwa S, Suzuki K, Makino F, Kajita T, Inoue T, Namba K, Watanabe T, Matsuura Y
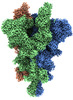
EMDB-33975: 
Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) with C480A mutation
Method: single particle / : Anzai I, Fujita J, Ono C, Kosaka Y, Miyamoto Y, Shichinohe S, Takada K, Torii S, Taguwa S, Suzuki K, Makino F, Kajita T, Inoue T, Namba K, Watanabe T, Matsuura Y

EMDB-35625: 
SARS-CoV-2 XBB.1 spike glycoprotein in complex with ACE2 (2-up state)
Method: single particle / : Anraku Y, Kita S, Yajima H, Sasaki J, Sasaki-Tabata K, Maenaka K, Hashiguchi T

EMDB-33593: 
Structure of the Anabaena PSI-monomer-IsiA supercomplex
Method: single particle / : Nagao R, Kato K, Hamaguchi T, Kawakami K, Yonekura K, Shen JR

EMDB-31526: 
human NTCP in complex with YN69083 Fab
Method: single particle / : Park JH, Iwamoto M, Yun JH, Uchikubo-Kamo T, Son D, Jin Z, Yoshida H, Ohki M, Ishimoto N, Mizutani K, Oshima M, Muramatsu M, Wakita T, Shirouzu M, Liu K, Uemura T, Nomura N, Iwata S, Watashi K, Tame JRH, Nishizawa T, Lee W, Park SY

PDB-7fci: 
human NTCP in complex with YN69083 Fab
Method: single particle / : Park JH, Iwamoto M, Yun JH, Uchikubo-Kamo T, Son D, Jin Z, Yoshida H, Ohki M, Ishimoto N, Mizutani K, Oshima M, Muramatsu M, Wakita T, Shirouzu M, Liu K, Uemura T, Nomura N, Iwata S, Watashi K, Tame JRH, Nishizawa T, Lee W, Park SY

EMDB-31455: 
Cryo-EM structure of a primordial cyanobacterial photosystem I
Method: single particle / : Kato K, Hamaguchi T, Nagao R, Kawakami K, Yonekura K, Shen JR

EMDB-31062: 
Structure of monomeric photosystem II
Method: single particle / : Yu H, Hamaguchi T, Nakajima Y, Kato K, kawakami K, Akita F, Yonekura K, Shen JR

PDB-7eda: 
Structure of monomeric photosystem II
Method: single particle / : Yu H, Hamaguchi T, Nakajima Y, Kato K, kawakami K, Akita F, Yonekura K, Shen JR

EMDB-30547: 
Cryo-EM Structure of PSII at 1.95 angstrom resolution
Method: single particle / : Kato K, Miyazaki N, Hamaguchi T, Nakajima Y, Akita F, Yonekura K, Shen JR

EMDB-30548: 
Cryo-EM Structure of PSII at 2.08 angstrom resolution
Method: single particle / : Kato K, Miyazaki N, Hamaguchi T, Nakajima Y, Akita F, Yonekura K, Shen JR

EMDB-30549: 
Cryo-EM Structure of PSII at 2.22 angstrom resolution
Method: single particle / : Kato K, Miyazaki N, Hamaguchi T, Nakajima Y, Akita F, Yonekura K, Shen JR

EMDB-30550: 
Cryo-EM Structure of PSII at 2.20 angstrom resolution
Method: single particle / : Kato K, Miyazaki N, Hamaguchi T, Nakajima Y, Akita F, Yonekura K, Shen JR

PDB-7d1t: 
Cryo-EM Structure of PSII at 1.95 angstrom resolution
Method: single particle / : Kato K, Miyazaki N, Hamaguchi T, Nakajima Y, Akita F, Yonekura K, Shen JR

PDB-7d1u: 
Cryo-EM Structure of PSII at 2.08 angstrom resolution
Method: single particle / : Kato K, Miyazaki N, Hamaguchi T, Nakajima Y, Akita F, Yonekura K, Shen JR

EMDB-12160: 
Cilia from MOT7 deletion mutant of Chlamydomonas
Method: subtomogram averaging / : Noga A, Kutomi O, Yamamoto R, Nakagiri Y, Imai H, Obbineni JM, Zimmermann N, Ishikawa T, Wakabayashi K, Kon T, Inaba K

EMDB-12161: 
Chlamydomonas cilia with MOT7-BCCP labeled
Method: subtomogram averaging / : Noga A, Kutomi O, Yamamoto R, Nakagiri Y, Imai H, Obbineni JM, Zimmermann N, Ishikawa T, Wakabayashi K, Kon T, Inaba K

EMDB-12162: 
Microtubule doublet structure from WT Chlamydomonas as a control for MOT7 mutants
Method: subtomogram averaging / : Obbineni JM, Kutomi O, Yamamoto R, Nakagiri Y, Imai H, Noga A, Zimmermann N, Ishikawa T, Wakabayashi K, Kon T, Inaba K

EMDB-9908: 
Structure of PSI-isiA supercomplex from Thermosynechococcus vulcanus
Method: single particle / : Akita F, Nagao R, Kato K, Shen JR, Miyazaki N

PDB-6k33: 
Structure of PSI-isiA supercomplex from Thermosynechococcus vulcanus
Method: single particle / : Akita F, Nagao R, Kato K, Shen JR, Miyazaki N

EMDB-10262: 
Yeast 80S ribosome stalled on SDD1 mRNA.
Method: single particle / : Tesina P, Buschauer R, Cheng J, Becker T, Beckmann R

EMDB-10315: 
The cryo-EM structure of SDD1-stalled collided trisome.
Method: single particle / : Tesina P, Buschauer R, Cheng J, Becker T, Beckmann R

PDB-6snt: 
Yeast 80S ribosome stalled on SDD1 mRNA.
Method: single particle / : Tesina P, Buschauer R, Cheng J, Becker T, Beckmann R

PDB-6sv4: 
The cryo-EM structure of SDD1-stalled collided trisome.
Method: single particle / : Tesina P, Buschauer R, Cheng J, Becker T, Beckmann R

EMDB-9530: 
Cryo-EM structure of the model post-termination complex (PoTC) in the rotated state
Method: single particle / : Iwakura N, Yokoyama T, Quaglia F, Mitsuoka K, Mio K, Shigematsu H, Shirouzu M, Kaji A, Kaji H

EMDB-9531: 
Cryo-EM structure of the model post-termination complex (PoTC) in the unrotated state
Method: single particle / : Iwakura N, Yokoyama T, Quaglia F, Mitsuoka K, Mio K, Shigematsu H, Shirouzu M, Kaji A, Kaji H

EMDB-9532: 
Cryo-EM structure of the model post-termination complex (PoTC) after the ribosome recycling reaction
Method: single particle / : Iwakura N, Yokoyama T, Quaglia F, Mitsuoka K, Mio K, Shigematsu H, Shirouzu M, Kaji A, Kaji H

EMDB-9533: 
Cryo-EM structure of the 70S ribosome from polysome
Method: single particle / : Iwakura N, Yokoyama T, Quaglia F, Mitsuoka K, Mio K, Shigematsu H, Shirouzu M, Kaji A, Kaji H

EMDB-8096: 
The Architecture of the Cytoplasmic Region of Type III Secretion Systems
Method: subtomogram averaging / : Makino F, Shen D, Kajimura N, Kawamoto A, Pissaridou P, Oswin H, Pain M, Murillo I, Namba K, Blocker AJ

EMDB-8121: 
The Architecture of the Cytoplasmic Region of Type III Secretion Systems
Method: subtomogram averaging / : Makino F, Blocker A

EMDB-8122: 
The Architecture of the Cytoplasmic Region of Type III Secretion Systems
Method: subtomogram averaging / : Makino F, Blocker A

EMDB-1915: 
Initial binding position of RRF on the post-termination complex
Method: single particle / : Yokoyama T, Shaikh TR, Iwakura N, Kaji H, Kaji A, Agrawal RK

EMDB-1916: 
Initial binding conformation of RRF on the post-termination complex
Method: single particle / : Yokoyama T, Shaikh TR, Iwakura N, Kaji H, Kaji A, Agrawal RK

EMDB-1917: 
Intermediate binding positions of RRF and EF-G on the post-termination complex
Method: single particle / : Yokoyama T, Shaikh TR, Iwakura N, Kaji H, Kaji A, Agrawal RK

EMDB-1918: 
Binding conformations and positions of RRF and EF-G during intermediate state of ribosome recycling
Method: single particle / : Yokoyama T, Shaikh TR, Iwakura N, Kaji H, Kaji A, Agrawal RK

PDB-3j0d: 
Models for the T. thermophilus ribosome recycling factor bound to the E. coli post-termination complex
Method: single particle / : Yokoyama T, Shaikh TR, Iwakura N, Kaji H, Kaji A, Agrawal RK

PDB-3j0e: 
Models for the T. thermophilus ribosome recycling factor and the E. coli elongation factor G bound to the E. coli post-termination complex
Method: single particle / : Yokoyama T, Shaikh TR, Iwakura N, Kaji H, Kaji A, Agrawal RK

EMDB-5254: 
WT Dam1 complex assembled into a ring around a microtubule
Method: single particle / : Ramey V, Wang HW, Nogales E

PDB-3iyo: 
Cryo-EM model of virion-sized HEV virion-sized capsid
Method: single particle / : Xing L, Mayazaki N, Li TC, Simons MN, Wall JS, Moore M, Wang CY, Takeda N, Wakita T, Miyamura T, Cheng RH

EMDB-5173: 
Cryo-EM structure of virion-sized hepatitis E virus-like particle
Method: single particle / : Xing L, Mayazaki N, Li TC, Simons MN, Wall JS, Moore M, Wang CY, Takeda N, Wakita T, Miyamura T, Cheng RH

EMDB-1369: 
Progression of the ribosome recycling factor through the ribosome dissociates the two ribosomal subunits.
Method: single particle / : Barat C, Datta PP, Raj VS, Sharma MR, Kaji H, Kaji A, Agrawal RK

EMDB-1370: 
Progression of the ribosome recycling factor through the ribosome dissociates the two ribosomal subunits.
Method: single particle / : Barat C, Datta PP, Raj VS, Sharma MR, Kaji H, Kaji A, Agrawal RK
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
