-Search query
-Search result
Showing 1 - 50 of 549 items for (author: julian & p)

EMDB-41711: 
Disulfide-stabilized HIV-1 CA hexamer in complex with PQBP1 Nt
Method: single particle / : Piacentini J, Pornillos O, Ganser-Pornillos BK

PDB-8ty6: 
Disulfide-stabilized HIV-1 CA hexamer in complex with PQBP1 Nt
Method: single particle / : Piacentini J, Pornillos O, Ganser-Pornillos BK

EMDB-17104: 
AApoAII amyloid fibril Morphology II (ex vivo)
Method: helical / : Andreotti G, Schmidt M, Faendrich M

EMDB-17105: 
AApoAII amyloid fibril Morphology I (ex vivo)
Method: helical / : Andreotti G, Schmidt M, Faendrich M

PDB-8oq4: 
AApoAII amyloid fibril Morphology II (ex vivo)
Method: helical / : Andreotti G, Schmidt M, Faendrich M

PDB-8oq5: 
AApoAII amyloid fibril Morphology I (ex vivo)
Method: helical / : Andreotti G, Schmidt M, Faendrich M

EMDB-41138: 
CryoEM structure of MFRV-VILP bound to IGF1Rzip
Method: single particle / : Kirk NS

EMDB-17736: 
ATTRV20I amyloid fibril from hereditary ATTR amloidosis
Method: helical / : Steinebrei M, Schmidt M, Faendrich M

EMDB-17737: 
ATTRG47E amyloid fibril from hereditary ATTR amloidosis
Method: helical / : Steinebrei M, Schmidt M, Faendrich M

EMDB-17738: 
ATTRV122I amyloid fibril from hereditary ATTR amloidosis
Method: helical / : Steinebrei M, Schmidt M, Faendrich M

PDB-8pke: 
ATTRV20I amyloid fibril from hereditary ATTR amloidosis
Method: helical / : Steinebrei M, Schmidt M, Faendrich M

PDB-8pkf: 
ATTRG47E amyloid fibril from hereditary ATTR amloidosis
Method: helical / : Steinebrei M, Schmidt M, Faendrich M

PDB-8pkg: 
ATTRV122I amyloid fibril from hereditary ATTR amloidosis
Method: helical / : Steinebrei M, Schmidt M, Faendrich M
EMDB-41649: 
P22 Mature Virion tail - C6 Localized Reconstruction
Method: single particle / : Iglesias S, Cingolani G, Feng-Hou C

EMDB-41651: 
In situ cryo-EM structure of bacteriophage P22 portal protein: head-to-tail protein complex at 3.0A resolution
Method: single particle / : Iglesias SM, Cingolani G, Feng-Hou C

EMDB-41819: 
In situ cryo-EM structure of bacteriophage P22 tailspike protein complex at 3.4A resolution
Method: single particle / : Iglesias SM, Feng-Hou C, Cingolani G

PDB-8tvr: 
In situ cryo-EM structure of bacteriophage P22 tail hub protein: tailspike protein complex at 2.8A resolution
Method: single particle / : Iglesias S, Cingolani G, Feng-Hou C

PDB-8tvu: 
In situ cryo-EM structure of bacteriophage P22 portal protein: head-to-tail protein complex at 3.0A resolution
Method: single particle / : Iglesias SM, Cingolani G, Feng-Hou C

PDB-8u1o: 
In situ cryo-EM structure of bacteriophage P22 tailspike protein complex at 3.4A resolution
Method: single particle / : Iglesias SM, Feng-Hou C, Cingolani G

EMDB-40678: 
Caspase-4/Pro-IL-18 complex
Method: single particle / : Pascal D, Dong Y, Wu H, Jon K

EMDB-41791: 
In situ cryo-EM structure of bacteriophage P22 gp1:gp4:gp5:gp10:gp9 N-term complex in conformation 1 at 3.2A resolution
Method: single particle / : Iglesias S, Feng-Hou C, Cingolani G

EMDB-41792: 
In situ cryo-EM structure of bacteriophage P22 gp1:gp5:gp4: gp10: gp9 N-term complex in conformation 2 at 3.1A resolution
Method: single particle / : Iglesias S, Feng-Hou C, Cingolani G

PDB-8u10: 
In situ cryo-EM structure of bacteriophage P22 gp1:gp4:gp5:gp10:gp9 N-term complex in conformation 1 at 3.2A resolution
Method: single particle / : Iglesias S, Feng-Hou C, Cingolani G

PDB-8u11: 
In situ cryo-EM structure of bacteriophage P22 gp1:gp5:gp4: gp10: gp9 N-term complex in conformation 2 at 3.1A resolution
Method: single particle / : Iglesias S, Feng-Hou C, Cingolani G

EMDB-29292: 
HIV-1 envelope trimer bound to one CD4 molecule on membranes
Method: subtomogram averaging / : Li W, Qin Z, Nand E, Grunst MW, Grover JR, Uchil PD, Mothes W

EMDB-29293: 
HIV-1 envelope trimer bound to two CD4 molecules on membranes
Method: subtomogram averaging / : Li W, Qin Z, Nand E, Grunst MW, Grover JR, Uchil PD, Mothes W

EMDB-29294: 
HIV-1 envelope trimer bound to three CD4 molecules on membranes
Method: subtomogram averaging / : Li W, Qin Z, Nand E, Grunst MW, Grover JR, Uchil PD, Mothes W

EMDB-29295: 
Consensus structure of HIV-1 envelope trimer bound to CD4 receptors on membranes
Method: subtomogram averaging / : Li W, Qin Z, Nand E, Grunst MW, Grover JR, Uchil PD, Mothes W

EMDB-41045: 
Unliganded HIV-1 Env on virion
Method: subtomogram averaging / : Li W, Qin Z, Nand E, Grunst MW, Grover JR, Uchil PD, Mothes W

EMDB-16458: 
Electron cryo-tomography and subtomogram averaging of cytoplasmic lattice filaments from mammalian oocytes
Method: subtomogram averaging / : Petrovic A, Bauerlein FJB, Jentoft IMA, Schuh M, Fernandez-Busnadiego R

EMDB-41464: 
H2 hemagglutinin (A/Singapore/1/1957) in complex with RBS-targeting 1-1-1F05
Method: single particle / : Yang YR, Han J, Perrett HR, Ward AB

EMDB-41465: 
H1 hemagglutinin (NC99) in complex with RBS-targeting Fab 1-1-1F05
Method: single particle / : Yang YR, Han J, Perrett HR, Ward AB

EMDB-41466: 
H2 hemagglutinin (A/Singapore/1/1957) in complex with RBS-targeting Fab 1-1-1E04
Method: single particle / : Yang YR, Han J, Perrett HR, Ward AB

EMDB-41467: 
H1 hemagglutinin (NC99) in complex with RBS-targeting Fab 1-1-1E04
Method: single particle / : Yang YR, Han J, Perrett HR, Ward AB

EMDB-41468: 
H2 hemagglutinin (A/Singapore/1/1957) in complex with RBS-targeting Fab 4-1-1E02
Method: single particle / : Yang YR, Han J, Perrett HR, Ward AB

EMDB-41469: 
H2 hemagglutinin (A/Singapore/1/1957) in complex with Sa-targeting Fab 4-1-1G03
Method: single particle / : Yang YR, Han J, Perrett HR, Ward AB

EMDB-41470: 
H2 hemagglutinin (A/Singapore/1/1957) in complex with medial-junction-targeting Fab 2-2-1G06
Method: single particle / : Yang YR, Han J, Perrett HR, Ward AB

EMDB-41471: 
H1 hemagglutinin (NC99) in complex with medial-junction-targeting Fab 2-2-1G06
Method: single particle / : Yang YR, Han J, Perrett HR, Ward AB

EMDB-41472: 
H2 hemagglutinin (A/Singapore/1/1957) in complex with polyclonal Fab, pAb_1
Method: single particle / : Yang YR, Han J, Perrett HR, Ward AB

EMDB-41473: 
H2 hemagglutinin (A/Singapore/1/1957) in complex with polyclonal Fab, pAb_2
Method: single particle / : Yang YR, Han J, Perrett HR, Ward AB

EMDB-41474: 
H2 hemagglutinin (A/Singapore/1/1957) in complex with polyclonal Fab, pAb_3
Method: single particle / : Yang YR, Han J, Perrett HR, Ward AB
EMDB-41523: 
Polyclonal immune complex of Fab binding the H2 HA from serum of subject 2-1 at week 4
Method: single particle / : Yang YR, Han J, Richey ST, Ward AB
EMDB-41524: 
Polyclonal immune complex of Fab binding the H2 HA from serum of subject 2-1 at week 16
Method: single particle / : Yang YR, Han J, Richey ST, Ward AB
EMDB-41525: 
Polyclonal immune complex of Fab binding the H2 HA from serum of subject 2-1 at week 20
Method: single particle / : Yang YR, Han J, Richey ST, Ward AB
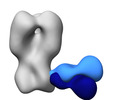
EMDB-41526: 
Polyclonal immune complex of Fab binding the H2 HA from serum of subject 2-2 at week 0
Method: single particle / : Yang YR, Han J, Richey ST, Ward AB
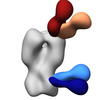
EMDB-41527: 
Polyclonal immune complex of Fab binding the H2 HA from serum of subject 2-2 at week 4
Method: single particle / : Yang YR, Han J, Richey ST, Ward AB

EMDB-41528: 
Polyclonal immune complex of Fab binding the H2 HA from serum of subject 2-2 week 16
Method: single particle / : Yang YR, Han J, Richey ST, Ward AB
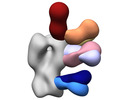
EMDB-41529: 
Polyclonal immune complex of Fab binding to H2 HA from serum of subject 2-2 at week 20
Method: single particle / : Yang YR, Han J, Richey ST, Ward AB
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
