-Search query
-Search result
Showing 1 - 50 of 243 items for (author: joseph & ap)

EMDB-41805: 
Cryo-EM structure of murine Thrombopoietin receptor ectodomain in complex with Tpo
Method: single particle / : Sarson-Lawrence KS, Hardy JM, Leis A, Babon JJ, Kershaw NJ

EMDB-18245: 
Plunge-frozen (control) map of beta-galactosidase
Method: single particle / : Esser TK, Boehning J, Bharat TAM, Rauschenbach S

EMDB-36766: 
Untilted (0 Degree Tilted) Single-Particle CryoEM Reconstruction of AAV2 Viral Capsid
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36767: 
10 Degree Tilted Single-Particle CryoEM Reconstruction of AAV2 Viral Capsid
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36768: 
20 Degree Tilted Single-Particle CryoEM Reconstruction of AAV2 Viral Capsid
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36769: 
30 Degree Tilted Single-Particle CryoEM Reconstruction of AAV2 Viral Capsid
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36770: 
40 Degree Tilted Single-Particle CryoEM Reconstruction of AAV2 Viral Capsid
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36771: 
50 Degree Tilted Single-Particle CryoEM Reconstruction of AAV2 Viral Capsid
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36772: 
60 Degree Tilted Single-Particle CryoEM Reconstruction of AAV2 Viral Capsid
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36807: 
Untilted (0 Degree Tilted) Single-Particle CryoEM Reconstruction of Apoferritin
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36809: 
10 Degree Tilted Single-Particle CryoEM Reconstruction of Apoferritin
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D
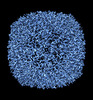
EMDB-36810: 
20 Degree Tilted Single-Particle CryoEM Reconstruction of Apoferritin
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36811: 
30 Degree Tilted Single-Particle CryoEM Reconstruction of Apoferritin
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36812: 
40 Degree Tilted Single-Particle CryoEM Reconstruction of Apoferritin
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36813: 
60 Degree Tilted Single-Particle CryoEM Reconstruction of Apoferritin
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D
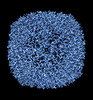
EMDB-36814: 
50 Degree Tilted Single-Particle CryoEM Reconstruction of Apoferritin
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36816: 
Untilted (0 Degree Tilted) Single-Particle CryoEM Reconstruction of DPS
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36817: 
10 Degree Tilted Single-Particle CryoEM Reconstruction of DPS
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36818: 
20 Degree Tilted Single-Particle CryoEM Reconstruction of DPS
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36819: 
30 Degree Tilted Single-Particle CryoEM Reconstruction of DPS
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36820: 
40 Degree Tilted Single-Particle CryoEM Reconstruction of DPS
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36821: 
50 Degree Tilted Single-Particle CryoEM Reconstruction of DPS
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-36822: 
60 Degree Tilted Single-Particle CryoEM Reconstruction of DPS
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-41230: 
Untilted (0 Degree Tilted) Single-Particle CryoEM Reconstruction of Apoferritin
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-41231: 
30 Degree Tilted Single-Particle CryoEM Reconstruction of Apoferritin
Method: single particle / : Aiyer S, Tan YZ, Lyumkis D

EMDB-41695: 
60 Degree Tilted Single-Particle CryoEM Reconstruction of RNA polymerase
Method: single particle / : Aiyer S, Shan Z, Oh J, Wang D, Tan YZ, Lyumkis D

EMDB-18244: 
ESIBD structure of beta-galactosidase
Method: single particle / : Esser T, Boehning J, Bharat TAM, Rauschenbach S

PDB-8q7y: 
ESIBD structure of beta-galactosidase
Method: single particle / : Esser T, Boehning J, Bharat TAM, Rauschenbach S

EMDB-17804: 
Bat-Hp-CoV Nsp1 and eIF1 bound to the human 40S small ribosomal subunit
Method: single particle / : Schubert K, Karousis ED, Ban I, Lapointe CP, Leibundgut M, Baeumlin E, Kummerant E, Scaiola A, Schoenhut T, Ziegelmueller J, Puglisi JD, Muehlemann O, Ban N

EMDB-17805: 
MERS-CoV Nsp1 bound to the human 43S pre-initiation complex
Method: single particle / : Schubert K, Karousis ED, Ban I, Lapointe CP, Leibundgut M, Baeumlin E, Kummerant E, Scaiola A, Schoenhut T, Ziegelmueller J, Puglisi JD, Muehlemann O, Ban N

PDB-8ppk: 
Bat-Hp-CoV Nsp1 and eIF1 bound to the human 40S small ribosomal subunit
Method: single particle / : Schubert K, Karousis ED, Ban I, Lapointe CP, Leibundgut M, Baeumlin E, Kummerant E, Scaiola A, Schoenhut T, Ziegelmueller J, Puglisi JD, Muehlemann O, Ban N

PDB-8ppl: 
MERS-CoV Nsp1 bound to the human 43S pre-initiation complex
Method: single particle / : Schubert K, Karousis ED, Ban I, Lapointe CP, Leibundgut M, Baeumlin E, Kummerant E, Scaiola A, Schoenhut T, Ziegelmueller J, Puglisi JD, Muehlemann O, Ban N

EMDB-40307: 
Sub-tomogram average of the RSV F pairs (with over-dosed particles removed) from the surface of native virions released from RSV-infected BEAS-2B cells cultured on EM grids collected via montage parallel array cryo- tomography (MPACT)
Method: subtomogram averaging / : Yang J, Wright ER

EMDB-40308: 
Sub-tomogram average of the RSV F pairs from the surface of native virions released from RSV-infected BEAS-2B cells cultured on EM grids collected via montage parallel array cryo-tomography (MPACT)
Method: subtomogram averaging / : Yang J, Wright ER

EMDB-15297: 
Mouse endoribonuclease Dicer
Method: single particle / : Zanova M, Zapletal D, Kubicek K, Stefl R, Pinkas M, Novacek J

EMDB-15298: 
Mouse endoribonuclease Dicer
Method: single particle / : Zanova M, Zapletal D, Kubicek K, Stefl R, Pinkas M, Novacek J

EMDB-15299: 
Mouse endoribonuclease Dicer
Method: single particle / : Zanova M, Zapletal D, Kubicek K, Stefl R, Pinkas M, Novacek J

EMDB-27943: 
9H2 Fab-poliovirus 1 complex
Method: single particle / : Charnesky AJ
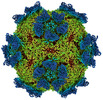
EMDB-27947: 
9H2 Fab-Sabin poliovirus 3 complex
Method: single particle / : Charnesky AJ

EMDB-27948: 
9H2 Fab-poliovirus 2 complex
Method: single particle / : Charnesky AJ
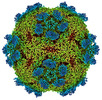
EMDB-27949: 
9H2 Fab-Sabin poliovirus 3 complex
Method: single particle / : Charnesky AJ
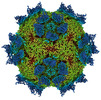
EMDB-27950: 
9H2 Fab-Sabin poliovirus 2 complex
Method: single particle / : Charnesky AJ
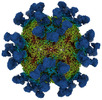
EMDB-27951: 
9H2 Fab-Sabin poliovirus 1 complex
Method: single particle / : Charnesky AJ

PDB-8e8l: 
9H2 Fab-poliovirus 1 complex
Method: single particle / : Charnesky AJ

PDB-8e8r: 
9H2 Fab-Sabin poliovirus 3 complex
Method: single particle / : Charnesky AJ

PDB-8e8s: 
9H2 Fab-poliovirus 2 complex
Method: single particle / : Charnesky AJ

PDB-8e8x: 
9H2 Fab-Sabin poliovirus 3 complex
Method: single particle / : Charnesky AJ

PDB-8e8y: 
9H2 Fab-Sabin poliovirus 2 complex
Method: single particle / : Charnesky AJ

PDB-8e8z: 
9H2 Fab-Sabin poliovirus 1 complex
Method: single particle / : Charnesky AJ
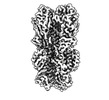
EMDB-40557: 
Cryo-EM structure of designed Influenza HA binder, HA_20, bound to Influenza HA (Strain: Iowa43)
Method: single particle / : Borst AJ, Bennett NR
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
