-Search query
-Search result
Showing 1 - 50 of 82 items for (author: josé & l & carrascosa)

EMDB-10911: 
Cryo-EM structure of T7 bacteriophage DNA translocation gp15 core protein intermediate assembly
Method: single particle / : Perez-Ruiz M, Pulido-Cid M, Luque-Ortega JR, Cuervo A, Carrascosa JL

EMDB-10912: 
Cryo-EM structure of T7 bacteriophage DNA translocation gp15-gp16 core complex intermediate assembly
Method: single particle / : Perez-Ruiz M, Pulido-Cid M, Luque-Ortega JR, Cuervo A, Carrascosa JL

PDB-6ysz: 
Cryo-EM structure of T7 bacteriophage DNA translocation gp15 core protein intermediate assembly
Method: single particle / : Perez-Ruiz M, Pulido-Cid M, Luque-Ortega JR, Cuervo A, Carrascosa JL

PDB-6yt5: 
Cryo-EM structure of T7 bacteriophage DNA translocation gp15-gp16 core complex intermediate assembly
Method: single particle / : Perez-Ruiz M, Pulido-Cid M, Luque-Ortega JR, Cuervo A, Carrascosa JL

EMDB-10010: 
Atomic structure of the Epstein-Barr portal, structure I
Method: single particle / : Machon C, Fabrega-Ferrer M, Zhou D, Cuervo A, Carrascosa JL, Stuart DI, Coll M

EMDB-10011: 
Atomic structure of the Epstein-Barr portal, structure II
Method: single particle / : Machon C, Fabrega-Ferrer M, Zhou D, Cuervo A, Carrascosa JL, Stuart DI, Coll M

PDB-6rvr: 
Atomic structure of the Epstein-Barr portal, structure I
Method: single particle / : Machon C, Fabrega-Ferrer M, Zhou D, Cuervo A, Carrascosa JL, Stuart DI, Coll M

PDB-6rvs: 
Atomic structure of the Epstein-Barr portal, structure II
Method: single particle / : Machon C, Fabrega-Ferrer M, Zhou D, Cuervo A, Carrascosa JL, Stuart DI, Coll M

EMDB-4667: 
Cryo-EM structure of T7 bacteriophage portal protein, 13mer, closed valve
Method: single particle / : Cuervo A, Fabrega-Ferrer M, Machon C, Fernandez FJ, Perez-Luque R, Pous J, Vega MC, Carrascosa JL, Coll M

EMDB-4669: 
Cryo-EM structure of T7 bacteriophage portal protein, 12mer, open valve
Method: single particle / : Fabrega-Ferrer M, Cuervo A, Machon C, Fernandez FJ, Perez-Luque R, Pous J, Vega MC, Carrascosa JL, Coll M

EMDB-4706: 
Cryo-EM structure of T7 bacteriophage fiberless tail complex
Method: single particle / : Cuervo A, Fabrega-Ferrer M, Machon C, Conesa JJ, Perez-Ruiz M, Coll M, Carrascosa JL

PDB-6qxm: 
Cryo-EM structure of T7 bacteriophage portal protein, 12mer, open valve
Method: single particle / : Fabrega-Ferrer M, Cuervo A, Machon C, Fernandez FJ, Perez-Luque R, Pous J, Vega MC, Carrascosa JL, Coll M

PDB-6r21: 
Cryo-EM structure of T7 bacteriophage fiberless tail complex
Method: single particle / : Cuervo A, Fabrega-Ferrer M, Machon C, Conesa JJ, Perez-Ruiz M, Coll M, Carrascosa JL

EMDB-3557: 
Centriolar Distal MT doublets in centrosome extracted from o.aries thymocytes
Method: subtomogram averaging / : Busselez J, Chichon FJ, Melero R, Carrascosa JL, Carazo JM

EMDB-3558: 
Centriolar Proximal MT triplet in centrosomes extracted from o.aries thymocytes
Method: subtomogram averaging / : Busselez J, Chichon FJ, Melero R, Carrascosa JL, Carazo JM

EMDB-3507: 
IBDV E1-1A population
Method: single particle / : Mata CP, Mertens J, Fontana J, Luque D, Allende-Ballestero C, Reguera D, Trus BL, Steven AC, Carrascosa JL, Caston JR

EMDB-3509: 
IBDV E1-1B population
Method: single particle / : Mata CP, Mertens J, Fontana J, Luque D, Allende-Ballestero C, Reguera D, Trus BL, Steven AC, Carrascosa JL, Caston JR

EMDB-3510: 
IBDV E1-2 population
Method: single particle / : Mata CP, Mertens J, Fontana J, Luque D, Allende-Ballestero C, Reguera D, Trus BL, Steven AC, Carrascosa JL, Caston JR

EMDB-3511: 
IBDV E1 population
Method: subtomogram averaging / : Mata CP, Mertens J, Fontana J, Luque D, Allende-Ballestero C, Reguera D, Trus BL, Steven AC, Carrascosa JL, Caston JR

EMDB-3512: 
IBDV E2 population
Method: subtomogram averaging / : Mata CP, Mertens J, Fontana J, Luque D, Allende-Ballestero C, Reguera D, Trus BL, Steven AC, Carrascosa JL, Caston JR

EMDB-3513: 
IBDV E3 population
Method: subtomogram averaging / : Mata CP, Mertens J, Fontana J, Luque D, Allende-Ballestero C, Reguera D, Trus BL, Steven AC, Carrascosa JL, Caston JR

EMDB-3514: 
IBDV E4 population
Method: subtomogram averaging / : Mata CP, Mertens J, Fontana J, Luque D, Allende-Ballestero C, Reguera D, Trus BL, Steven AC, Carrascosa JL, Caston JR

EMDB-3515: 
IBDV E5 population
Method: subtomogram averaging / : Mata CP, Mertens J, Fontana J, Luque D, Allende-Ballestero C, Reguera D, Trus BL, Steven AC, Carrascosa JL, Caston JR

EMDB-3516: 
IBDV E6 population
Method: subtomogram averaging / : Mata CP, Mertens J, Fontana J, Luque D, Allende-Ballestero C, Reguera D, Trus BL, Steven AC, Carrascosa JL, Caston JR

EMDB-3619: 
RnQV1-W1118 empty capsid
Method: single particle / : Mata CP, Luque D, Gomez-Blanco J, Rodriguez JM, Suzuki N, Ghabrial SA, Carrascosa JL, Trus BL, Caston JR

PDB-5nd1: 
Viral evolution results in multiple, surface-allocated enzymatic activities in a fungal double-stranded RNA virus
Method: single particle / : Mata CP, Luque D, Gomez Blanco J, Rodriguez JM, Suzuki N, Ghabrial SA, Carrascosa JL, Trus BL, Caston JR

EMDB-5996: 
Electron cryo-microscopy of chimeric EGFP-VP2 capsid
Method: single particle / : Pascual E, Mata CP, Gomez-Blanco J, Carrascosa JL, Caston JR

EMDB-5997: 
Electron cryo-microscopy of chimeric HT-VP2-466 capsid
Method: single particle / : Pascual E, Mata CP, Gomez-Blanco J, Carrascosa JL, Caston JR

EMDB-2717: 
Electron cryo-microscopy of T7 bacteriophage tail after DNA ejection
Method: single particle / : Gonzalez-Garcia VA, Pulido-Cid M, Garcia-Doval C, van Raaij MJ, Martin-Benito J, Cuervo A, Carrascosa JL

EMDB-2573: 
SA11 Rotavirus non-trypsinized Triple Layered Particle
Method: single particle / : Rodriguez JM, Chichon FJ, Martin-Forero E, Gonzalez-Camacho F, Carrascosa JL, Caston JR, Luque D

EMDB-2574: 
SA11 Rotavirus trypsinized Triple Layered Particle
Method: single particle / : Rodriguez JM, Chichon FJ, Martin-Forero E, Gonzalez-Camacho F, Carrascosa JL, Caston JR, Luque D

EMDB-2575: 
SA11 Rotavirus non-trypsinized Triple Layered Particle grown in absence of leupetin
Method: single particle / : Rodriguez JM, Chichon FJ, Martin-Forero E, Gonzalez-Camacho F, Carrascosa JL, Caston JR, Luque D

EMDB-2576: 
OSU Rotavirus non-trypsinized Triple Layered Particle
Method: single particle / : Rodriguez JM, Chichon FJ, Martin-Forero E, Gonzalez-Camacho F, Carrascosa JL, Caston JR, Luque D

EMDB-2577: 
OSU Rotavirus trypsinized Triple Layered Particle
Method: single particle / : Rodriguez JM, Chichon FJ, Martin-Forero E, Gonzalez-Camacho F, Carrascosa JL, Caston JR, Luque D
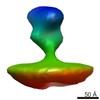
EMDB-2578: 
Class 1 averaged spike from SA11 Rotavirus non-trypsinized Triple Layered Particle
Method: subtomogram averaging / : Rodriguez JM, Chichon FJ, Martin-Forero E, Gonzalez-Camacho F, Carrascosa JL, Caston JR, Luque D

EMDB-2579: 
Class 2 averaged spike from SA11 Rotavirus non-trypsinized Triple Layered Particle
Method: subtomogram averaging / : Rodriguez JM, Chichon FJ, Martin-Forero E, Gonzalez-Camacho F, Carrascosa JL, Caston JR, Luque D
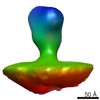
EMDB-2580: 
Class 1 averaged spike from SA11 Rotavirus trypsinized Triple Layered Particle
Method: subtomogram averaging / : Rodriguez JM, Chichon FJ, Martin-Forero E, Gonzalez-Camacho F, Carrascosa JL, Caston JR, Luque D

EMDB-5600: 
Penicillium Chrysogenum Virus (PcV) capsid structure
Method: single particle / : Luque D, Gomez-Blanco J, Garriga D, Brilot A, Gonzalez JM, Havens WH, Carrascosa JL, Trus BL, Verdaguer N, Grigorieff N, Ghabrial SA, Caston JR

PDB-3j3i: 
Penicillium chrysogenum virus (PcV) capsid structure
Method: single particle / : Luque D, Gomez-Blanco J, Garriga D, Brilot A, Gonzalez JM, Havens WH, Carrascosa JL, Trus BL, Verdaguer N, Grigorieff N, Ghabrial SA, Caston JR

EMDB-5713: 
Electron microscopy of negatively-stained gp12 tubular protein of T7 bacteriophage
Method: single particle / : Cuervo A, Pulido-Cid M, Chagoyen M, Arranz R, Gonzalez-Garcia VA, Garcia-Doval C, Caston JR, Valpuesta JM, van Raaij MJ, Martin-Benito J, Carrascosa JL

EMDB-5689: 
Cryo-electron microscopy of T7 tail complex
Method: single particle / : Cuervo A, Pulido-Cid M, Chagoyen M, Arranz R, Gonzalez-Garcia VA, Garcia-Doval C, Caston JR, Valpuesta JM, van Raaij MJ, Martin-Benito J, Carrascosa JL

EMDB-5690: 
Cryo-electron microscopy of T7 tail complex formed by gp8, gp11, and gp12 proteins
Method: single particle / : Cuervo A, Pulido-Cid M, Chagoyen M, Arranz R, Gonzalez-Garcia VA, Garcia-Doval C, Caston JR, Valpuesta JM, van Raaij MJ, Martin-Benito J, Carrascosa JL

PDB-3j4a: 
Structure of gp8 connector protein
Method: single particle / : Cuervo A, Pulido-Cid M, Chagoyen M, Arranz R, Gonzalez-Garcia VA, Garcia-Doval C, Caston JR, Valpuesta JM, van Raaij MJ, Martin-Benito J, Carrascosa JL

PDB-3j4b: 
Structure of T7 gatekeeper protein (gp11)
Method: single particle / : Cuervo A, Pulido-Cid M, Chagoyen M, Arranz R, Gonzalez-Garcia VA, Garcia-Doval C, Caston JR, Valpuesta JM, van Raaij MJ, Martin-Benito J, Carrascosa JL
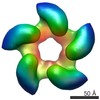
EMDB-2355: 
Negative staining three dimensional reconstruction of bacteriophage T7 large terminase
Method: single particle / : Dauden MI, Martin-Benito J, Sanchez-Ferrero JC, Pulido-Cid M, Valpuesta JM, Carrascosa JL

EMDB-2356: 
Negative staining three-dimensional reconstruction of bacteriophage T7 connector/terminase complex
Method: single particle / : Dauden MI, Martin-Benito J, Sanchez-Ferrero JC, Pulido-Cid M, Valpuesta JM, Carrascosa JL

PDB-4bij: 
Threading model of T7 large terminase
Method: single particle / : Dauden MI, Martin-Benito J, Sanchez-Ferrero JC, Pulido-Cid M, Valpuesta JM, Carrascosa JL

PDB-4bil: 
Threading model of the T7 large terminase within the gp8gp19 complex
Method: single particle / : Dauden MI, Martin-Benito J, Sanchez-Ferrero JC, Pulido-Cid M, Valpuesta JM, Carrascosa JL

EMDB-2205: 
Cryo-electron microscopy reconstruction of the helical part of influenza A virus ribonucleoprotein isolated from virions.
Method: helical / : Arranz R, Coloma R, Chichon FJ, Conesa JJ, Carrascosa JL, Valpuesta JM, Ortin J, Martin-Benito J

EMDB-2206: 
Negative stained electron microscopy reconstruction of the loop of influenza A virus ribonucleoprotein isolated from virions.
Method: single particle / : Arranz R, Coloma R, Chichon FJ, Conesa JJ, Carrascosa JL, Valpuesta JM, Ortin J, Martin-Benito J
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
