-Search query
-Search result
Showing 1 - 50 of 92 items for (author: johnson & gt)
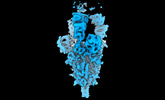
EMDB-14885: 
OMI-42 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE GLYCOPROTEIN
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-14886: 
OMI-38 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE RBD (local refinement)
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-14887: 
OMI-2 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE GLYCOPROTEIN
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI
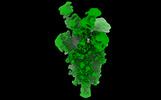
EMDB-14910: 
OMI-38 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-7zr7: 
OMI-42 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE GLYCOPROTEIN
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-7zr8: 
OMI-38 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE RBD (local refinement)
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-7zr9: 
OMI-2 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE GLYCOPROTEIN
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-7zrc: 
OMI-38 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-13857: 
Beta049 fab in complex with SARS-CoV2 beta-Spike glycoprotein, The Beta mAb response underscores the antigenic distance to other SARS-CoV-2 variants
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-13868: 
Beta-50 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-13869: 
COVOX-222 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-13870: 
Beta-43 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-13871: 
Beta-26 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-13872: 
Beta-32 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-13873: 
Beta-53 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-13874: 
Beta-44 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-13875: 
Beta-06 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-7q6e: 
Beta049 fab in complex with SARS-CoV2 beta-Spike glycoprotein, The Beta mAb response underscores the antigenic distance to other SARS-CoV-2 variants
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-7q9f: 
Beta-50 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-7q9g: 
COVOX-222 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-7q9i: 
Beta-43 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-7q9j: 
Beta-26 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-7q9k: 
Beta-32 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-7q9m: 
Beta-53 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-7q9p: 
Beta-06 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-12774: 
Cryo-electron tomogram of mature RSV CANC CLPs
Method: electron tomography / : Obr M, Ricana CL, Nikulin N, Feathers JPR, Klanschnig M, Thader A, Johnson MC, Vogt VM, Schur FKM, Dick RA

EMDB-12485: 
Structure of the mature RSV CA lattice: T=1 CA icosahedron
Method: single particle / : Obr M, Ricana CL, Nikulin N, Feathers JPR, Klanschnig M, Thader A, Johnson MC, Vogt VM, Schur FKM, Dick RA

EMDB-12486: 
Structure of the mature RSV CA lattice: T=3 CA icosahedron
Method: single particle / : Obr M, Ricana CL, Nikulin N, Feathers JPR, Klanschnig M, Thader A, Johnson MC, Vogt VM, Schur FKM, Dick RA

EMDB-12487: 
Structure of the mature RSV CA lattice: hexamer derived from tubes (C2-symmetric)
Method: subtomogram averaging / : Obr M, Ricana CL, Nikulin N, Feathers JPR, Klanschnig M, Thader A, Johnson MC, Vogt VM, Schur FKM, Dick RA

EMDB-12488: 
Structure of the mature RSV CA lattice: pentamer derived from polyhedral VLPs
Method: subtomogram averaging / : Obr M, Ricana CL, Nikulin N, Feathers JPR, Klanschnig M, Thader A, Johnson MC, Vogt VM, Schur FKM, Dick RA

EMDB-12489: 
Structure of the mature RSV CA lattice: hexamer with 3 adjacent pentamers (C3 symmetric)
Method: subtomogram averaging / : Obr M, Ricana CL, Nikulin N, Feathers JPR, Klanschnig M, Thader A, Johnson MC, Vogt VM, Schur FKM, Dick RA

EMDB-12490: 
Structure of the mature RSV CA lattice: hexamer with 2 adjacent pentamers (C2 symmetric)
Method: subtomogram averaging / : Obr M, Ricana CL, Nikulin N, Feathers JPR, Klanschnig M, Thader A, Johnson MC, Vogt VM, Schur FKM, Dick RA

EMDB-12491: 
Structure of the mature RSV CA lattice: Group I, pentamer-hexamer interface, class 1
Method: subtomogram averaging / : Obr M, Ricana CL, Nikulin N, Feathers JPR, Klanschnig M, Thader A, Johnson MC, Vogt VM, Schur FKM, Dick RA

EMDB-12492: 
Structure of the mature RSV CA lattice: Group I, pentamer-hexamer interface, class 1"2
Method: subtomogram averaging / : Obr M, Ricana CL, Nikulin N, Feathers JPR, Klanschnig M, Thader A, Johnson MC, Vogt VM, Schur FKM, Dick RA

EMDB-12493: 
Structure of the mature RSV CA lattice: Group I, pentamer-hexamer interface, class 1"6
Method: subtomogram averaging / : Obr M, Ricana CL, Nikulin N, Feathers JPR, Klanschnig M, Thader A, Johnson MC, Vogt VM, Schur FKM, Dick RA

EMDB-12494: 
Structure of the mature RSV CA lattice: Group I, pentamer-pentamer interface, class 1'1
Method: subtomogram averaging / : Obr M, Ricana CL, Nikulin N, Feathers JPR, Klanschnig M, Thader A, Johnson MC, Vogt VM, Schur FKM, Dick RA

EMDB-12495: 
Structure of the mature RSV CA lattice: Group II, hexamer-hexamer interface, class 6
Method: subtomogram averaging / : Obr M, Ricana CL, Nikulin N, Feathers JPR, Klanschnig M, Thader A, Johnson MC, Vogt VM, Schur FKM, Dick RA

EMDB-12496: 
Structure of the mature RSV CA lattice: Group II, hexamer-hexamer interface, class 2'6
Method: subtomogram averaging / : Obr M, Ricana CL, Nikulin N, Feathers JPR, Klanschnig M, Thader A, Johnson MC, Vogt VM, Schur FKM, Dick RA

EMDB-12497: 
Structure of the mature RSV CA lattice: Group III, hexamer-hexamer interface, class 3'3
Method: subtomogram averaging / : Obr M, Ricana CL, Nikulin N, Feathers JPR, Klanschnig M, Thader A, Johnson MC, Vogt VM, Schur FKM, Dick RA

EMDB-12498: 
Structure of the mature RSV CA lattice: Group III, hexamer-hexamer interface, class 3'4
Method: subtomogram averaging / : Obr M, Ricana CL, Nikulin N, Feathers JPR, Klanschnig M, Thader A, Johnson MC, Vogt VM, Schur FKM, Dick RA

EMDB-12499: 
Structure of the mature RSV CA lattice: Group III, hexamer-hexamer interface, class 3'5
Method: subtomogram averaging / : Obr M, Ricana CL, Nikulin N, Feathers JPR, Klanschnig M, Thader A, Johnson MC, Vogt VM, Schur FKM, Dick RA

EMDB-12500: 
Structure of the mature RSV CA lattice: Group III, hexamer-hexamer interface, class 4'4
Method: subtomogram averaging / : Obr M, Ricana CL, Nikulin N, Feathers JPR, Klanschnig M, Thader A, Johnson MC, Vogt VM, Schur FKM, Dick RA
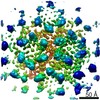
EMDB-12501: 
Structure of the mature RSV CA lattice: Group III, hexamer-hexamer interface, class 4'5
Method: subtomogram averaging / : Obr M, Ricana CL, Nikulin N, Feathers JPR, Klanschnig M, Thader A, Johnson MC, Vogt VM, Schur FKM, Dick RA

EMDB-12502: 
Structure of the mature RSV CA lattice: Group III, hexamer-hexamer interface, class 5'5
Method: subtomogram averaging / : Obr M, Ricana CL, Nikulin N, Feathers JPR, Klanschnig M, Thader A, Johnson MC, Vogt VM, Schur FKM, Dick RA

EMDB-12503: 
Structure of the mature RSV CA lattice: Group IV, hexamer-hexamer interface, class 3'Alpha
Method: subtomogram averaging / : Obr M, Ricana CL, Nikulin N, Feathers JPR, Klanschnig M, Thader A, Johnson MC, Vogt VM, Schur FKM, Dick RA

EMDB-12504: 
Structure of the mature RSV CA lattice: Group IV, hexamer-hexamer interface, class 3'Beta
Method: subtomogram averaging / : Obr M, Ricana CL, Nikulin N, Feathers JPR, Klanschnig M, Thader A, Johnson MC, Vogt VM, Schur FKM, Dick RA

EMDB-12505: 
Structure of the mature RSV CA lattice: Group IV, hexamer-hexamer interface, class 3'Gamma
Method: subtomogram averaging / : Obr M, Ricana CL, Nikulin N, Feathers JPR, Klanschnig M, Thader A, Johnson MC, Vogt VM, Schur FKM, Dick RA

EMDB-12506: 
Structure of the mature RSV CA lattice: Group IV, hexamer-hexamer interface, class 4'Alpha
Method: subtomogram averaging / : Obr M, Ricana CL, Nikulin N, Feathers JPR, Klanschnig M, Thader A, Johnson MC, Vogt VM, Schur FKM, Dick RA

EMDB-12507: 
Structure of the mature RSV CA lattice: Group IV, hexamer-hexamer interface, class 4'Beta
Method: subtomogram averaging / : Obr M, Ricana CL, Nikulin N, Feathers JPR, Klanschnig M, Thader A, Johnson MC, Vogt VM, Schur FKM, Dick RA

EMDB-12508: 
Structure of the mature RSV CA lattice: Group IV, hexamer-hexamer interface, class 4'Gamma
Method: subtomogram averaging / : Obr M, Ricana CL, Nikulin N, Feathers JPR, Klanschnig M, Thader A, Johnson MC, Vogt VM, Schur FKM, Dick RA
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
