-Search query
-Search result
Showing all 47 items for (author: johnson & cm)

EMDB-29068: 
Cryo-EM structure of the GR-Hsp90-FKBP52 complex
Method: single particle / : Noddings CM, Agard DA

EMDB-29069: 
Cryo-EM structure of the GR-Hsp90-FKBP51 complex
Method: single particle / : Noddings CM, Agard DA

EMDB-41153: 
Integrin alpha-v beta-8 in complex with minibinder B8_BP_dsulf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

EMDB-41154: 
Integrin alpha-v beta-6 in complex with minibinder B6_BP_dslf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D
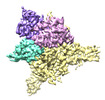
EMDB-27898: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with 2-oxohexadecyl-CoA
Method: single particle / : Johnson ZL, Wasilko DJ, Ammirati M, Chang JS, Han S, Wu H
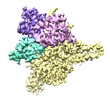
EMDB-27899: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with CoA and palmitoyl-LPA
Method: single particle / : Wasilko DJ, Johnson ZL, Ammirati M, Chang JS, Han S, Wu H

EMDB-26507: 
SARS-CoV-2 spike in complex with Multivalent miniprotein inhibitor FUS231-P24 (2RBDs open)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-26508: 
SARS-CoV-2 spike in complex with multivalent miniprotein inhibitor FUS231-P24 (3RBDs open)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-26509: 
SARS-CoV-2 spike in complex with multivalent miniprotein inhibitor FUS31-G10 (2RBDs open)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-26510: 
SARS-CoV-2 spike in complex with multivalent miniprotein inhibitor FUS31-G10 (3RBDs open)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-26511: 
SARS-CoV-2 spike in complex with AHB2-2GS-SB175 (local refinement of the RBD and AHB2)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-26512: 
SARS-CoV-2 spike in complex with AHB2-2GS-SB175
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-26429: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the neutralizing antibody Fab fragment, C1520
Method: single particle / : Barnes CO

EMDB-26430: 
Structure of the SARS-CoV-2 NTD in complex with C1520, local refinement
Method: single particle / : Barnes CO

EMDB-26431: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the neutralizing antibody Fab fragment, C1717
Method: single particle / : Barnes CO

EMDB-26432: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the neutralizing antibody Fab fragment, C1791
Method: single particle / : Barnes CO

EMDB-25574: 
SARS-CoV-2 S-RBD + Fab 54042-4
Method: single particle / : Johnson NV, Mclellan JS

EMDB-23004: 
The GR-Maturation Complex: Glucocorticoid Receptor in complex with Hsp90 and co-chaperone p23
Method: single particle / : Noddings CM, Wang YR, Agard DA

EMDB-23005: 
Maltose Binding Protein in complex with Hsp90 and co-chaperone p23
Method: single particle / : Noddings CM, Wang YR, Agard DA

EMDB-23006: 
Complex of Hsp90 and co-chaperone p23
Method: single particle / : Noddings CM, Wang YR, Agard DA

EMDB-23050: 
Atomic cryoEM structure of Hsp90-Hsp70-Hop-GR
Method: single particle / : Wang RY, Noddings CM, Kirschke E, Myasnikov A, Johnson JL, Agard DA

EMDB-23051: 
Focused map of the Hsp90-Hsp70-Hop-GR client-loading complex: Hsp90-CTDs:Hop-DP2:GR-Helix1
Method: single particle / : Wang RY, Agard DA

EMDB-23053: 
Focused map of the Hsp90-Hsp70-Hop-GR complex: Hsp90A-NTD-MD:Hsp70C-NBD
Method: single particle / : Wang RY, Agard DA

EMDB-23054: 
Focused map of the Hsp90-Hsp70-Hop-GR complex: Hsp90B-NTD-MD:Hsp70D-NBD
Method: single particle / : Wang RY, Agard DA

EMDB-23055: 
Focused map of the Hsp90-Hsp70-Hop-GR complex: Hsp70D-NBD:Hop-TPR2A
Method: single particle / : Wang RY, Agard DA

EMDB-23056: 
Focused map of the Hsp90-Hsp70-Hop-GR complex: Hsp90B-NTD-MD:Hsp70D-NBD:Hop-TPR2A-TPR2B-DP2
Method: single particle / : Wang RY, Agard DA

EMDB-23156: 
SARS-CoV 2 Spike Protein bound to LY-CoV555
Method: single particle / : Goldsmith JA, McLellan JS

EMDB-22829: 
Human Tom70 in complex with SARS CoV2 Orf9b
Method: single particle / : QCRG Structural Biology Consortium

EMDB-10132: 
cryo-EM structure of mTORC1 bound to PRAS40-fused active RagA/C GTPases
Method: single particle / : Anandapadamanaban M, Berndt A, Masson GR, Perisic O, Williams RL

EMDB-10133: 
cryo-EM structure of mTORC1 bound to active RagA/C GTPases
Method: single particle / : Anandapadamanaban M, Berndt A, Masson GR, Perisic O, Williams RL

EMDB-4371: 
Cryo-EM map of the FCHSD2 F-BAR domain + SH3-1
Method: single particle / : Almeida-Souza L, Frank R, Garcia-Nafria J, Colussi A, Gunawardana N, Johnson CM, Yu M, Howard G, Andrews B, Vallis Y, McMahon HT

EMDB-3668: 
Closed dimer (C2) of human ATM (Ataxia telangiectasia mutated)
Method: single particle / : Baretic D, Johnson CM, Santhanam B, Truman CM, Kouba T, Fersht AR, Phillips C, Williams RL, Pollard HK, Fisher DI

EMDB-3669: 
Closed dimer (C1) of human ATM (Ataxia telangiectasia mutated)
Method: single particle / : Baretic D, Pollard HK, Fisher DI, Johnson CM, Santhanam B, Truman CM, Kouba T, Fersht AR, Phillips C, Williams RL

EMDB-3670: 
Closed dimer Pincer-FATKIN of human ATM (Ataxia telangiectasia mutated)
Method: single particle / : Baretic D, Pollard HK, Fisher DI, Johnson CM, Santhanam B, Truman CM, Kouba T, Fersht AR, Phillips C, Williams RL

EMDB-3671: 
Open dimer of human ATM (Ataxia telangiectasia mutated)
Method: single particle / : Baretic D, Pollard HK, Fisher DI, Johnson CM, Santhanam B, Truman CM, Kouba T, Fersht AR, Phillips C

EMDB-3672: 
Open protomer of human ATM (Ataxia telangiectasia mutated)
Method: single particle / : Baretic D, Pollard HK, Fisher DI, Johnson CM, Santhanam B, Truman CM, Kouba T, Fersht AR, Phillips C, Williams RL

EMDB-3673: 
Open protomer Pincer-FATKIN of human ATM (Ataxia telangiectasia mutated)
Method: single particle / : Baretic D, Pollard HK, Fisher DI, Johnson CM, Santhanam B, Truman CM, Kouba T, Fersht AR, Phillips C
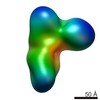
EMDB-8235: 
Negative stain structure of Vps15/Vps34 complex
Method: single particle / : Kirsten ML, Zhang L, Ohashi Y, Perisic O, Williams RL, Sachse C

EMDB-3329: 
Cryo electron microscopy of a complex of Tor and Lst8
Method: single particle / : Baretic D, Berndt A, Ohashi Y, Johnson CM, Williams RL

EMDB-3334: 
Cryo electron microscopy of a complex of Tor-323RFP and Lst8
Method: single particle / : Baretic D, Berndt A, Ohashi Y, Johnson CM, Williams RL

EMDB-3335: 
Cryo electron microscopy of a complex of Tor-1190RFP and Lst8
Method: single particle / : Baretic D, Berndt A, Ohashi Y, Johnson CM, Williams RL

EMDB-3336: 
Cryo electron microscopy of a complex of Tor-1175RFP and Lst8
Method: single particle / : Baretic D, Berndt A, Ohashi Y, Johnson CM, Williams RL

EMDB-2477: 
Negative stain electron microscopy structure of native human Presenilin 1 (PS1) complex
Method: single particle / : Li Y, Lu S, Tsai CJ, Bohm C, Qamar S, Dodd RB, Meadows W, Jeon A, McLeod A, Chen F, Arimon M, Berezovska O, Hyman BT, Tomita T, Iwatsubod T, Johnsof CM, Farrer L, Schmitt-Ulms G, Fraser P, St George-Hyslop P

EMDB-2478: 
Negative stain electron microscopy structure of compound E-bound human Presenilin 1 (PS1) complex
Method: single particle / : Li Y, Lu S, Tsai CJ, Bohm C, Qamar S, Dodd RB, Meadows W, Jeon A, McLeod A, Chen F, Arimon M, Berezovska O, Hyman BT, Tomita T, Iwatsubod T, Johnsof CM, Farrer L, Schmitt-Ulms G, Fraser P, St George-Hyslop P

EMDB-5584: 
Life in the extremes: atomic structure of Sulfolobus Turreted Icosahedral Virus
Method: single particle / : Veesler D, Ng TS, Sendamarai AK, Eilers BJ, Lawrence CM, Lok SM, Young MJ, Johnson JE, Fu CY

EMDB-5149: 
P22 Procapsid Shell
Method: single particle / : Parent KN, Khayat R, Tu LH, Suhanovsky MM, Coritnes JR, Teschke CM, Johnson JE, Baker TS

EMDB-5150: 
P22 Heat Expanded Head
Method: single particle / : Parent KN, Khayat R, Tu LH, Suhanovsky MM, Cortines JR, Teschke CM, Johnson JE, Baker TS
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
