-Search query
-Search result
Showing all 38 items for (author: joazeiro & cap)

EMDB-14156: 
Cryo-EM reconstruction of the Bacillus subtilis MutS2-collided disome complex (MutS2 conf.1; Leading 70S)
Method: single particle / : Filbeck S, Pfeffer S

EMDB-14157: 
Composite reconstruction of the Bacillus subtilis collided disome (Leading 70S)
Method: single particle / : Filbeck S, Pfeffer S

EMDB-14158: 
Composite reconstruction of the Bacillus subtilis collided disome (Collided 70S)
Method: single particle / : Filbeck S, Pfeffer S

EMDB-14159: 
Cryo-EM reconstruction of the Bacillus subtilis MutS2-collided disome complex (MutS2 conf.2; Leading 70S)
Method: single particle / : Filbeck S, Pfeffer S

EMDB-14160: 
Cryo-EM reconstruction of the Bacillus subtilis MutS2-collided disome complex (Leading 70S)
Method: single particle / : Filbeck S, Pfeffer S

EMDB-14161: 
Cryo-EM reconstruction of the Bacillus subtilis MutS2-collided disome complex (Collided 70S)
Method: single particle / : Filbeck S, Pfeffer S

EMDB-14162: 
Cryo-EM reconstruction of the Bacillus subtilis collided disome (Leading 70S)
Method: single particle / : Filbeck S, Pfeffer S

EMDB-14163: 
Cryo-EM reconstruction of the Bacillus subtilis collided disome (Leading 30S)
Method: single particle / : Filbeck S, Pfeffer S

EMDB-14164: 
Cryo-EM reconstruction of the Bacillus subtilis collided disome (Collided 70S)
Method: single particle / : Filbeck S, Pfeffer S

EMDB-14165: 
Cryo-EM reconstruction of the Bacillus subtilis collided disome (Collided 30S)
Method: single particle / : Filbeck S, Pfeffer S

EMDB-14166: 
Cryo-EM reconstruction of Bacillus subtilis obstructed 50S subunit co-purified with MutS2
Method: single particle / : Filbeck S, Pfeffer S

EMDB-11862: 
Structure of the bacterial RQC complex (Decoding State)
Method: single particle / : Filbeck S, Pfeffer S

EMDB-11864: 
Structure of the bacterial RQC complex (Translocating State)
Method: single particle / : Filbeck S, Pfeffer S

EMDB-4799: 
B.subtilis 50S subunit-nascent chain-tRNA complex
Method: single particle / : Lytvynenko I, Paternoga H, Thrun A, Balke A, Mueller T, Nagler K, Chiang CH, Tsaprailis G, Anders S, Maupin-Furlow JA, Bischofs I, Spahn CMT, Joazeiro CAP

EMDB-2797: 
single-particle cryo reconstruction of the Large Ribosomal subunit-associated protein Quality Control (RQC) complex
Method: single particle / : Lyumkis D, Oliveira dos Passos D, Tahara EB, Webb K, Bennett EJ, Vinterbo S, Potter CS, Carragher B, Joazeiro CAP

EMDB-2798: 
single-particle cryo reconstruction of the Large Ribosomal subunit-associated protein Quality Control (RQC) complex in the context of Tae2 deletion
Method: single particle / : Lyumkis D, Oliveira dos Passos D, Tahara EB, Webb K, Bennett EJ, Vinterbo S, Potter CS, Carragher B, Joazeiro CAP

EMDB-2248: 
negative stain single-particle reconstruction of conformation I of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP
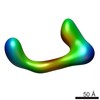
EMDB-2249: 
negative stain single-particle reconstruction of conformation II of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP

EMDB-2250: 
negative stain single-particle reconstruction of conformation III of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP
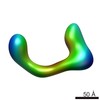
EMDB-2251: 
negative stain single-particle reconstruction of conformation IV of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP
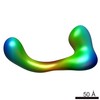
EMDB-2252: 
negative stain single-particle reconstruction of conformation V of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP
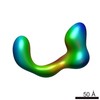
EMDB-2253: 
negative stain single-particle reconstruction of conformation VI of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP
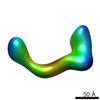
EMDB-2254: 
negative stain single-particle reconstruction of conformation VII of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP
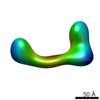
EMDB-2255: 
negative stain single-particle reconstruction of conformation VIII of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP

EMDB-2256: 
negative stain single-particle reconstruction of conformation IX of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP
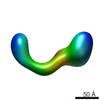
EMDB-2257: 
negative stain single-particle reconstruction of conformation X of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP

EMDB-2258: 
negative stain single-particle reconstruction of conformation XI of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP

EMDB-2259: 
negative stain single-particle reconstruction of conformation XII of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP

EMDB-2260: 
negative stain single-particle reconstruction of conformation XIII of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP

EMDB-2261: 
negative stain single-particle reconstruction of conformation XIV of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP

EMDB-2262: 
negative stain single-particle reconstruction of conformation XV of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP

EMDB-2263: 
negative stain single-particle reconstruction of conformation XVI of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP
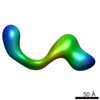
EMDB-2264: 
negative stain single-particle reconstruction of conformation XVII of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP

EMDB-2265: 
negative stain single-particle reconstruction of conformation XVIII of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP

EMDB-2266: 
negative stain single-particle reconstruction of conformation XIX of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP

EMDB-2267: 
negative stain single-particle reconstruction of conformation XX of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP

EMDB-2268: 
negative stain single-particle reconstruction of conformation XXI of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP
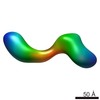
EMDB-2269: 
negative stain single-particle reconstruction of conformation XXII of the Ltn1 E3 ubiquitin Ligase
Method: single particle / : Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, Petroski MD, Lima CD, Potter CS, Carragher B, Joazeiro CAP
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
