-Search query
-Search result
Showing 1 - 50 of 283 items for (author: jin & aj)

EMDB-41637: 
Asymmetric cryoEM reconstruction of Mayaro virus
Method: single particle / : Chmielewski D, Kaelber J, Jin J, Weaver S, Auguste AJ, Chiu W

EMDB-41907: 
Computationally Designed, Expandable O4 Octahedral Handshake Nanocage
Method: single particle / : Weidle C, Borst A
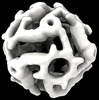
EMDB-42031: 
Computational Designed Nanocage O43_129_+8
Method: single particle / : Weidle C, Kibler RD

EMDB-28979: 
Cryo-EM structure of Chikungunya virus asymmetric unit
Method: single particle / : Su GC, Chmielewsk D, Kaelber J, Pintilie G, Chen M, Jin J, Auguste A, Chiu W

EMDB-41096: 
Cryo-electron tomography of Chikungunya virus pentamer structure
Method: subtomogram averaging / : Chmielewsk D, Su GC, Kaelber J, Pintilie G, Chen M, Jin J, Auguste A, Chiu W

EMDB-41631: 
Cryo-EM structure of Chikungunya virus with asymmetric reconstruction
Method: single particle / : Su GC, Chmielewsk D, Kaelber J, Pintilie G, Chen M, Jin J, Auguste A, Chiu W

EMDB-43318: 
Twistless helix 12 repeat ring design R12B
Method: single particle / : Calise SJ, Kollman JM
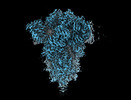
EMDB-18807: 
SD1-2 fab in complex with SARS-COV-2 BA.12.1 Spike Glycoprotein.
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI
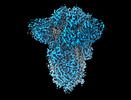
EMDB-18808: 
SD1-3 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-8r1c: 
SD1-2 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-8r1d: 
SD1-3 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-29974: 
Cryo-EM structure of synthetic tetrameric building block sC4
Method: single particle / : Redler RL, Huddy TF, Hsia Y, Baker D, Ekiert D, Bhabha G

EMDB-41364: 
CryoEM Structure of a Computationally Designed T3 Tetrahedral Nanocage
Method: single particle / : Weidle C, Borst AJ

EMDB-42906: 
Computational Designed Nanocage O43_129
Method: single particle / : Weidle C, Kibler RD

EMDB-42944: 
Computational Designed Nanocage O43_129_+4
Method: single particle / : Carr KD, Weidle C, Borst AJ

PDB-8gel: 
Cryo-EM structure of synthetic tetrameric building block sC4
Method: single particle / : Redler RL, Huddy TF, Hsia Y, Baker D, Ekiert D, Bhabha G

PDB-8tl7: 
CryoEM Structure of a Computationally Designed T3 Tetrahedral Nanocage
Method: single particle / : Weidle C, Borst AJ

PDB-8v3b: 
Computational Designed Nanocage O43_129_+4
Method: single particle / : Carr KD, Weidle C, Borst AJ
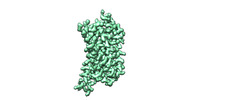
EMDB-43732: 
momSalB bound Kappa Opioid Receptor in complex Gi1
Method: single particle / : Fay JF, Che T

EMDB-43733: 
GR89,696 bound Kappa Opioid Receptor in complex with Gz
Method: single particle / : Fay JF, Che T

EMDB-43734: 
GR89,696 bound Kappa Opioid Receptor in complex with gustducin
Method: single particle / : Fay JF, Che T

EMDB-16680: 
BA.4/5-5 FAB IN COMPLEX WITH SARS-COV-2 BA.4 SPIKE GLYCOPROTEIN
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI, Fry EE

PDB-8cin: 
BA.4/5-5 FAB IN COMPLEX WITH SARS-COV-2 BA.4 SPIKE GLYCOPROTEIN
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI, Fry EE

EMDB-41042: 
S. enterica WbaP in a styrene maleic acid liponanoparticle
Method: single particle / : Dodge GJ, Imperiali B

EMDB-34981: 
Cryo-EM structure of human NTCP-myr-preS1-YN9048Fab complex
Method: single particle / : Asami J, Shimizu T, Ohto U

EMDB-34982: 
Cryo-EM structure of human NTCP-myr-preS1-YN9016Fab complex
Method: single particle / : Asami J, Shimizu T, Ohto U

PDB-8hrx: 
Cryo-EM structure of human NTCP-myr-preS1-YN9048Fab complex
Method: single particle / : Asami J, Shimizu T, Ohto U

PDB-8hry: 
Cryo-EM structure of human NTCP-myr-preS1-YN9016Fab complex
Method: single particle / : Asami J, Shimizu T, Ohto U

EMDB-29452: 
Structure Of Respiratory Syncytial Virus Polymerase with Novel Non-Nucleoside Inhibitor
Method: single particle / : Yu X, Abeywickrema P, Bonneux B, Behera I, Jacoby E, Fung A, Adhikary S, Bhaumik A, Carbajo RJ, Bruyn SD, Miller R, Patrick A, Pham Q, Piassek M, Verheyen N, Shareef A, Sutto-Ortiz P, Ysebaert N, Vlijmen HV, Jonckers THM, Herschke F, McLellan JS, Decroly E, Fearns R, Grosse S, Roymans D, Sharma S, Rigaux P, Jin Z

PDB-8fu3: 
Structure Of Respiratory Syncytial Virus Polymerase with Novel Non-Nucleoside Inhibitor
Method: single particle / : Yu X, Abeywickrema P, Bonneux B, Behera I, Jacoby E, Fung A, Adhikary S, Bhaumik A, Carbajo RJ, Bruyn SD, Miller R, Patrick A, Pham Q, Piassek M, Verheyen N, Shareef A, Sutto-Ortiz P, Ysebaert N, Vlijmen HV, Jonckers THM, Herschke F, McLellan JS, Decroly E, Fearns R, Grosse S, Roymans D, Sharma S, Rigaux P, Jin Z
EMDB-41039: 
MERS-CoV Nsp1 protein bound to the Human 40S Ribosomal subunit
Method: single particle / : Devarkar SC, Xiong Y

EMDB-41063: 
MERS-CoV Nsp1 protein binds the human 40S ribosome - Consensus Reconstruction
Method: single particle / : Devarkar SC, Xiong Y
EMDB-41064: 
MERS-CoV Nsp1 protein bound to the human 40S ribosome: Focus Refined 40S Head Map
Method: single particle / : Devarkar SC, Xiong Y
EMDB-41065: 
MERS-CoV Nsp1 protein bound to human 40S ribosome: 40S Body Focus Refined Map
Method: single particle / : Devarkar SC, Xiong Y

PDB-8t4s: 
MERS-CoV Nsp1 protein bound to the Human 40S Ribosomal subunit
Method: single particle / : Devarkar SC, Xiong Y

EMDB-41876: 
momSalB bound Kappa Opioid Receptor in complex with GoA
Method: single particle / : Fay JF, Che T

EMDB-16416: 
HB3VAR03 apo headstructure (PfEMP1 A) complexed with EPCR
Method: single particle / : Raghavan SSR, Lavstsen T, Wang KT

PDB-8c44: 
HB3VAR03 apo headstructure (PfEMP1 A) complexed with EPCR
Method: single particle / : Raghavan SSR, Lavstsen T, Wang KT

EMDB-16415: 
HB3VAR03 apo headstructure (PfEMP1 A)
Method: single particle / : Raghavan SSR, Lavstsen T, Wang KT

PDB-8c3y: 
HB3VAR03 apo headstructure (PfEMP1 A)
Method: single particle / : Raghavan SSR, Lavstsen T, Wang KT

EMDB-33650: 
SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7
Method: single particle / : Chia WN, Tan CW, Tan AWK, Young B, Starr TN, Lopez E, Fibriansah G, Barr J, Cheng S, Yeoh AYY, Yap WC, Lim BL, Ng TS, Sia WR, Zhu F, Chen S, Zhang J, Greaney AJ, Chen M, Au GG, Paradkar P, Peiris M, Chung AW, Bloom JD, Lye D, Lok SM, Wang LF

EMDB-33651: 
SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7 (focused refinement on Fab-RBD interface)
Method: single particle / : Chia WN, Tan CW, Tan AWK, Young B, Starr TN, Lopez E, Fibriansah G, Barr J, Cheng S, Yeoh AYY, Yap WC, Lim BL, Ng TS, Sia WR, Zhu F, Chen S, Zhang J, Greaney AJ, Chen M, Au GG, Paradkar P, Peiris M, Chung AW, Bloom JD, Lye D, Lok SM, Wang LF

EMDB-29930: 
T. cruzi topoisomerase II alpha bound to dsDNA and the covalent inhibitor CT1
Method: single particle / : Schenk A, Deniston C, Noeske J

PDB-8gcc: 
T. cruzi topoisomerase II alpha bound to dsDNA and the covalent inhibitor CT1
Method: single particle / : Schenk A, Deniston C, Noeske J

EMDB-29915: 
CryoEM map of a de novo designed octahedral nanocage with programmable volume; design cage_O4_34
Method: single particle / : Kibler RD, Borst AJ

EMDB-40070: 
Cryo-EM map of synthetic cage_O3_10 reconstructed without symmetry (C1)
Method: single particle / : Coudray N, Redler R, Hsia Y, Huddy TF, Baker D, Ekiert D, Bhabha G

EMDB-40071: 
Cryo-EM map of synthetic cage_O3_10 reconstructed with O symmetry
Method: single particle / : Coudray N, Redler R, Hsia Y, Huddy TF, Baker D, Ekiert D, Bhabha G

EMDB-40073: 
Cryo-EM map of synthetic cage_T3_5 reconstructed without symmetry (C1), with 1 monomer missing (class 3.0)
Method: single particle / : Coudray N, Redler R, Huddy TF, Hsia Y, Baker D, Ekiert D, Bhabha G

EMDB-40074: 
Cryo-EM map of synthetic cage_T3_5 reconstructed with T symmetry
Method: single particle / : Coudray N, Redler R, Huddy TF, Hsia Y, Baker D, Ekiert D, Bhabha G
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
