-Search query
-Search result
Showing 1 - 50 of 431 items for (author: jeon & h)

EMDB-41356: 
Structure of the C. elegans TMC-2 complex
Method: single particle / : Clark S, Jeong H, Goehring A, Posert R, Gouaux E

PDB-8tkp: 
Structure of the C. elegans TMC-2 complex
Method: single particle / : Clark S, Jeong H, Goehring A, Posert R, Gouaux E

EMDB-43091: 
hGBP1 conformer on the bacterial outer membrane
Method: subtomogram averaging / : Zhu S, MacMicking J

EMDB-43153: 
hGBP1 conformer on the bacterial outer membrane
Method: subtomogram averaging / : Zhu S, MacMicking J
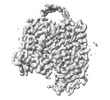
EMDB-35377: 
Cryo-EM structure of GPR156 of GPR156-miniGo-scFv16 complex (local refine)
Method: single particle / : Shin J, Park J, Cho Y

EMDB-35378: 
Cryo-EM structure of miniGo-scFv16 of GPR156-miniGo-scFv16 complex (local refine)
Method: single particle / : Shin J, Park J, Cho Y

EMDB-35380: 
Cryo-EM structure of GPR156-miniGo-scFv16 complex
Method: single particle / : Shin J, Park J, Cho Y

EMDB-35382: 
Cryo-EM structure of GPR156A/B of G-protein free GPR156 (local refine)
Method: single particle / : Shin J, Park J, Cho Y

EMDB-35389: 
Cryo-EM structure of GPR156C/D of G-protein free GPR156 (local refine)
Method: single particle / : Shin J, Park J, Cho Y

EMDB-35390: 
Cryo-EM structure of G-protein free GPR156
Method: single particle / : Shin J, Park J, Cho Y

PDB-8ieb: 
Cryo-EM structure of GPR156 of GPR156-miniGo-scFv16 complex (local refine)
Method: single particle / : Shin J, Park J, Cho Y

PDB-8iec: 
Cryo-EM structure of miniGo-scFv16 of GPR156-miniGo-scFv16 complex (local refine)
Method: single particle / : Shin J, Park J, Cho Y

PDB-8ied: 
Cryo-EM structure of GPR156-miniGo-scFv16 complex
Method: single particle / : Shin J, Park J, Cho Y

PDB-8iei: 
Cryo-EM structure of GPR156A/B of G-protein free GPR156 (local refine)
Method: single particle / : Shin J, Park J, Cho Y

PDB-8iep: 
Cryo-EM structure of GPR156C/D of G-protein free GPR156 (local refine)
Method: single particle / : Shin J, Park J, Cho Y

PDB-8ieq: 
Cryo-EM structure of G-protein free GPR156
Method: single particle / : Shin J, Park J, Cho Y

EMDB-34155: 
Designed pH-responsive P22 VLP
Method: single particle / : Kim KJ, Kim G, Bae JH, Song JJ, Kim HS

PDB-8gn5: 
Designed pH-responsive P22 VLP
Method: single particle / : Kim KJ, Kim G, Bae JH, Song JJ, Kim HS
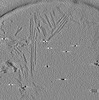
EMDB-28752: 
Cryo-electron tomography of wild type a-synuclein preformed fibrils
Method: electron tomography / : Jiang J, Boparai N, Dai W, Kim YS
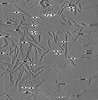
EMDB-28753: 
Cryo-electron tomography of S42Y a-synuclein preformed fibrils
Method: electron tomography / : Jiang J, Boparai N, Dai W, Kim YS

EMDB-34022: 
Cryo-EM structure of human topoisomerase II beta in complex with DNA and etoposide
Method: single particle / : Naganuma M, Ehara H, Kim D, Nakagawa R, Cong A, Bu H, Jeong J, Jang J, Schellenberg MJ, Bunch H, Sekine S

PDB-7yq8: 
Cryo-EM structure of human topoisomerase II beta in complex with DNA and etoposide
Method: single particle / : Naganuma M, Ehara H, Kim D, Nakagawa R, Cong A, Bu H, Jeong J, Jang J, Schellenberg MJ, Bunch H, Sekine S

EMDB-35904: 
AtSLAC1 8D mutant in closed state
Method: single particle / : Lee Y, Lee S

EMDB-35920: 
AtSLAC1 in open state
Method: single particle / : Lee Y, Lee S

EMDB-34303: 
AtSLAC1 6D mutant in closed state
Method: single particle / : Lee Y, Lee S

EMDB-34304: 
AtSLAC1 6D mutant in open state
Method: single particle / : Lee Y, Lee S

EMDB-40088: 
HIV-1 Env subtype C CZA97.12 SOSIP.664 in complex with 3BNC117 Fab
Method: single particle / : Ozorowski G, Lee JH, Ward AB

PDB-8gje: 
HIV-1 Env subtype C CZA97.12 SOSIP.664 in complex with 3BNC117 Fab
Method: single particle / : Ozorowski G, Lee JH, Ward AB

EMDB-29530: 
SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-29531: 
SARS-CoV-2 BQ.1.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-40240: 
SARS-CoV-2 BN.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

PDB-8fxb: 
SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

PDB-8fxc: 
SARS-CoV-2 BQ.1.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

PDB-8s9g: 
SARS-CoV-2 BN.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-34394: 
Structure of a TRP channel-G protein complex, additional map 2
Method: single particle / : Won J, Jeong H, Lee HH

EMDB-34395: 
Structure of a TRP channel-G protein complex, additional map 1
Method: single particle / : Won J, Jeong H, Lee HH

EMDB-34396: 
Structure of a TRP channel-G protein complex, Class2, additional map 1
Method: single particle / : Won J, Jeong H, Lee HH

EMDB-33274: 
Human Cx36/GJD2 (N-terminal deletion mutant) gap junction channel in soybean lipids (D6 symmetry)
Method: single particle / : Lee SN, Cho HJ, Jeong H, Ryu B, Lee HJ, Lee HH, Woo JS

PDB-7xl8: 
Human Cx36/GJD2 (N-terminal deletion mutant) gap junction channel in soybean lipids (D6 symmetry)
Method: single particle / : Lee SN, Cho HJ, Jeong H, Ryu B, Lee HJ, Lee HH, Woo JS

EMDB-40253: 
Truncated Braf/Mek/14-3-3 complex
Method: single particle / : Eck MJ, Jeon H, Park E, Rawson S

EMDB-33275: 
Human Cx36/GJD2 (N-terminal deletion mutant) gap junction channel in soybean lipids (C1 symmetry)
Method: single particle / : Lee SN, Cho HJ, Jeong H, Ryu B, Lee HJ, Lee HH, Woo JS

EMDB-33402: 
Formate dehydrogenase (FDH) from Methylobacterium extorquens AM1 (MeFDH1)
Method: single particle / : Park J, Heo YY, Roh SH, Lee HH

EMDB-33872: 
Human Cytosolic 10-formyltetrahydrofolate dehydrogenase and Gossypol complex
Method: single particle / : Han CW, Lee HN, Jeong MS, Jang SB

PDB-7yjj: 
Human Cytosolic 10-formyltetrahydrofolate dehydrogenase and Gossypol complex
Method: single particle / : Han CW, Lee HN, Jeong MS, Jang SB

EMDB-27428: 
Cryo-EM structure of a RAS/RAF complex (state 1)
Method: single particle / : Eck MJ, Jeon H, Park E, Rawson S

EMDB-27429: 
Cryo-EM structure of a RAS/RAF complex (state 2)
Method: single particle / : Eck MJ, Jeon H, Park E, Rawson S
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search







 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
