-Search query
-Search result
Showing 1 - 50 of 58 items for (author: jamie & h & d & cate)

EMDB-29449: 
E. coli 70S ribosome with an improved MS2 Tag inserted in H98 (Composite Map)
Method: single particle / : Nissley AJ, Cate JHD

EMDB-29483: 
E. coli 70S ribosome with an improved MS2 tag inserted in H98 (70S map)
Method: single particle / : Nissley AJ, Cate JHD

EMDB-29484: 
E. coli 70S ribosome with an improved MS2 tag inserted in H98 (50S focused map)
Method: single particle / : Nissley AJ, Cate JHD

EMDB-29485: 
E. coli 70S ribosome with an improved MS2 tag inserted in H98 (30S focused map)
Method: single particle / : Nissley AJ, Cate JHD

PDB-8fto: 
E. coli 70S ribosome with an improved MS2 tag inserted in H98
Method: single particle / : Nissley AJ, Cate JHD
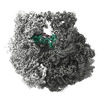
EMDB-28254: 
Composite 70S ribosome structure for "Atomistic simulations of the E. coli ribosome provide selection criteria for translationally active substrates
Method: single particle / : Watson ZL, Cate JHD
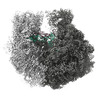
EMDB-28255: 
70S map for: "Atomistic simulations of the E. coli ribosome provide selection criteria for translationally active substrates"
Method: single particle / : Watson ZL, Cate JHD

EMDB-28256: 
30S-focused map for: "Atomistic simulations of the E. coli ribosome provide selection criteria for translationally active substrates"
Method: single particle / : Watson ZL, Cate JHD
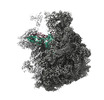
EMDB-28257: 
50S-focused map for: "Atomistic simulations of the E. coli ribosome provide selection criteria for translationally active substrates"
Method: single particle / : Watson ZL, Cate JHD

PDB-8emm: 
Composite 70S ribosome structure for "Atomistic simulations of the E. coli ribosome provide selection criteria for translationally active substrates
Method: single particle / : Watson ZL, Cate JHD

EMDB-29790: 
30S focus refined map of WT E.coli ribosome complexed with A-site ortho-aminobenzoic acid charged tRNA-Phe
Method: single particle / : Majumdar C, Cate JHD

EMDB-29786: 
Structure of WT E.coli 70S ribosome complexed with mRNA, P-site fMet-NH-tRNAfMet and A-site ortho-aminobenzoic acid charged NH-tRNAPhe
Method: single particle / : Majumdar C, Cate JHD

EMDB-29788: 
Structure of WT E.coli ribosome 50S subunit with complexed with mRNA, P-site fMet-NH-tRNAfMet and A-site 3-aminopyridine-4-carboxylic acid charged NH-tRNAPhe
Method: single particle / : Majumdar C, Cate JHD

PDB-8g6w: 
Structure of WT E.coli 70S ribosome complexed with mRNA, P-site fMet-NH-tRNAfMet and A-site ortho-aminobenzoic acid charged NH-tRNAPhe
Method: single particle / : Majumdar C, Cate JHD

PDB-8g6y: 
Structure of WT E.coli ribosome 50S subunit with complexed with mRNA, P-site fMet-NH-tRNAfMet and A-site 3-aminopyridine-4-carboxylic acid charged NH-tRNAPhe
Method: single particle / : Majumdar C, Cate JHD

EMDB-29787: 
Structure of WT E.coli ribosome 50S subunit with complexed with mRNA, P-site fMet-NH-tRNAfMet and A-site meta-aminobenzoic acid charged NH-tRNAPhe
Method: single particle / : Majumdar C, Cate JHD

PDB-8g6x: 
Structure of WT E.coli ribosome 50S subunit with complexed with mRNA, P-site fMet-NH-tRNAfMet and A-site meta-aminobenzoic acid charged NH-tRNAPhe
Method: single particle / : Majumdar C, Cate JHD
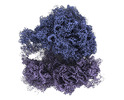
EMDB-28165: 
E. coli 70S ribosome with A-loop mutations U2554C and U2555C (Composite Map)
Method: single particle / : Nissley AJ, Penev PI, Watson ZL, Banfield JF, Cate JHD

EMDB-28218: 
E. coli 70S ribosome with A-loop mutations U2554C and U2555C (50S Focus Refinement)
Method: single particle / : Nissley AJ, Penev PI, Watson ZL, Banfield JF, Cate JHD

EMDB-28229: 
E. coli 70S ribosome with A-loop mutations U2554C and U2555C (30S Focus Refinement)
Method: single particle / : Nissley AJ, Penev PI, Watson ZL, Banfield JF, Cate JHD

EMDB-28230: 
E. coli 70S ribosome with A-loop mutations U2554C and U2555C: A-site tRNA class 2
Method: single particle / : Nissley AJ, Penev PI, Watson ZL, Banfield JF, Cate JHD

PDB-8eiu: 
E. coli 70S ribosome with A-loop mutations U2554C and U2555C
Method: single particle / : Nissley AJ, Penev PI, Watson ZL, Banfield JF, Cate JHD

EMDB-22086: 
Intermediate ribosome nascent chain complex during PF846 stalled translation termination
Method: single particle / : Li W, Cate J, Ward RF, Chang S

EMDB-22085: 
Selectively stalling of translation termination by a drug-like compound
Method: single particle / : Li W, Cate J, Ward RF, Chang S

PDB-6xa1: 
Structure of a drug-like compound stalled human translation termination complex
Method: single particle / : Li W, Cate J

EMDB-22586: 
Structure of the Bacterial Ribosome at 2 Angstrom Resolution (composite structure)
Method: single particle / : Watson ZL, Ward FR

EMDB-22607: 
Structure of the Bacterial Ribosome at 2 Angstrom Resolution (70S ribosome)
Method: single particle / : Watson ZL, Ward FR, Meheust R, Ad O, Schepartz A, Banfield JF, Cate JHD

EMDB-22614: 
Structure of the Bacterial Ribosome at 2 Angstrom Resolution (50S focused refinement)
Method: single particle / : Watson ZL, Ward FR, Meheust R, Ad O, Schepartz A, Banfield JF, Cate JHD

EMDB-22632: 
Structure of the Bacterial Ribosome at 2 Angstrom Resolution (30S focused refinement)
Method: single particle / : Watson ZL, Ward FR, Meheust R, Ad O, Schepartz A, Banfield JF, Cate JHD

EMDB-22635: 
Structure of the Bacterial Ribosome at 2 Angstrom Resolution (30S head domain focused refinement)
Method: single particle / : Watson ZL, Ward FR, Meheust R, Ad O, Schepartz A, Banfield JF, Cate JHD

EMDB-22636: 
Structure of the Bacterial Ribosome at 2 Angstrom Resolution (30S platform domain focused refinement)
Method: single particle / : Watson ZL, Ward FR, Meheust R, Ad O, Schepartz A, Banfield JF, Cate JHD

EMDB-22637: 
Structure of the Bacterial Ribosome at 2 Angstrom Resolution (50S central protuberance focused refinement)
Method: single particle / : Watson ZL, Ward FR, Meheust R, Ad O, Schepartz A, Banfield JF, Cate JHD

PDB-7k00: 
Structure of the Bacterial Ribosome at 2 Angstrom Resolution
Method: single particle / : Watson ZL, Ward FR, Meheust R, Ad O, Schepartz A, Banfield JF, Cate JHD

EMDB-20853: 
P7A7 ribosome large subunit
Method: single particle / : Watson ZL, Ward FR, Cate JHD

EMDB-20854: 
E. coli large ribosomal subunit
Method: single particle / : Watson ZL, Ward FR, Cate JHD

EMDB-20638: 
E. coli 50S with phazolicin (PHZ) bound in exit tunnel
Method: single particle / : Watson ZL, Cate JHD

PDB-6u48: 
E. coli 50S with phazolicin (PHZ) bound in exit tunnel
Method: single particle / : Watson ZL, Cate JHD, Polikanov YS, Severinov K

PDB-6ole: 
Human ribosome nascent chain complex (CDH1-RNC) stalled by a drug-like molecule with AP and PE tRNAs
Method: single particle / : Li W, Cate JHD

PDB-6olf: 
Human ribosome nascent chain complex (CDH1-RNC) stalled by a drug-like molecule with AA and PE tRNAs
Method: single particle / : Li W, Cate JHD

PDB-6olg: 
Human ribosome nascent chain complex stalled by a drug-like small molecule (CDH1_RNC with PP tRNA)
Method: single particle / : Li W, Cate JHD

PDB-6oli: 
Structure of human ribosome nascent chain complex selectively stalled by a drug-like small molecule (USO1-RNC)
Method: single particle / : Li W, Cate JHD

PDB-6olz: 
Human ribosome nascent chain complex (PCSK9-RNC) stalled by a drug-like molecule with PP tRNA
Method: single particle / : Li W, Cate JHD

PDB-6om0: 
Human ribosome nascent chain complex (PCSK9-RNC) stalled by a drug-like molecule with AP and PE tRNAs
Method: single particle / : Li W, Cate JHD

PDB-6om7: 
Human ribosome nascent chain complex (PCSK9-RNC) stalled by a drug-like small molecule with AA and PE tRNAs
Method: single particle / : Li W, Cate JHD

EMDB-0526: 
Structure of drug-like molecule stalled USO1 ribosome nascent chain complex (human)
Method: single particle / : Li W, Cate JHD

EMDB-20134: 
Structure of a drug-like molecule stalled PCSK9 ribosome nascent chain complex (PCSK9-RNC) with AP tRNA and PE tRNA (sample prepared with a short incubation time)
Method: single particle / : Li W, Cate JHD

EMDB-20135: 
Structure of a drug-like molecule stalled PCSK9 ribosome nascent chain complex (PCSK9-RNC) with PP tRNA (sample prepared with a short incubation time)
Method: single particle / : Li W, Cate JHD

EMDB-0596: 
Structure of a drug-like molecule stalled PCSK9 ribosome nascent chain complex (PCSK9-RNC) with AP tRNA and PE tRNA
Method: single particle / : Li W, Cate JHD

EMDB-0597: 
Structure of a drug-like molecule stalled ribosome nascent chain complex (PCSK9-RNC) in rotated state with AA tRAN and PE tRNA
Method: single particle / : Li W, Cate JHD

EMDB-0598: 
Structure of a drug-like molecule stalled ribosome nascent chain complex (PCSK9_RNC) in non-rotated state with PP tRNA
Method: single particle / : Li W, Cate JHD
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
