-Search query
-Search result
Showing 1 - 50 of 637 items for (author: james & z & chen)

EMDB-41672: 
ELIC5 with Propylamine in spNW15 nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL

EMDB-41673: 
ELIC with Propylamine in spNW15 nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL

PDB-8twv: 
ELIC5 with Propylamine in spNW15 nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL

PDB-8twz: 
ELIC with Propylamine in spNW15 nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL
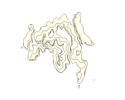
EMDB-19184: 
Late alpha-Synuclein fibril structure from liquid-liquid phase separations.
Method: helical / : De Simone A, Barritt JD, Chen S, Cascella R, Cecchi C, Bigi A, Jarvis JA, Chiti F, Dobson CM, Fusco G

PDB-8ri9: 
Late alpha-Synuclein fibril structure from liquid-liquid phase separations.
Method: helical / : De Simone A, Barritt JD, Chen S, Cascella R, Cecchi C, Bigi A, Jarvis JA, Chiti F, Dobson CM, Fusco G

EMDB-38095: 
MRE-269 bound Prostacyclin Receptor G protein complex
Method: single particle / : Wang JJ, Jin S, Zhang H, Xu Y, Hu W, Jiang Y, Chen C, Wang DW, Xu HE, Wu C
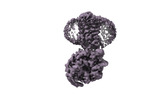
EMDB-38096: 
Treprostinil bound Prostacyclin Receptor G protein complex
Method: single particle / : Wang JJ, Jin S, Zhang H, Xu Y, Hu W, Jiang Y, Chen C, Wang DW, Xu HE, Wu C

PDB-8x79: 
MRE-269 bound Prostacyclin Receptor G protein complex
Method: single particle / : Wang JJ, Jin S, Zhang H, Xu Y, Hu W, Jiang Y, Chen C, Wang DW, Xu HE, Wu C

PDB-8x7a: 
Treprostinil bound Prostacyclin Receptor G protein complex
Method: single particle / : Wang JJ, Jin S, Zhang H, Xu Y, Hu W, Jiang Y, Chen C, Wang DW, Xu HE, Wu C

EMDB-43542: 
ELIC5 with cysteamine in 2:1:1 POPC:POPE:POPG nanodisc in open conformation
Method: single particle / : Petroff II JT, Deng Z, Rau MJ, Fitzpatrick JAJ, Yuan P, Cheng WWL

PDB-8vuw: 
ELIC5 with cysteamine in 2:1:1 POPC:POPE:POPG nanodisc in open conformation
Method: single particle / : Petroff II JT, Deng Z, Rau MJ, Fitzpatrick JAJ, Yuan P, Cheng WWL

EMDB-41830: 
Lipidated recombinant apolipoprotein E4
Method: single particle / : Strickland MR, Rau M, Summers B, Basore K, Wulf II J, Jiang H, Chen Y, Ulrich JD, Randolph GJ, Zhang R, Fitzpatrick JAJ, Cashikar AG, Holtzman DM

EMDB-41831: 
Gradient-fixed lipidated recombinant apolipoprotein E4
Method: single particle / : Strickland MR, Rau M, Summers B, Basore K, Wulf II J, Jiang H, Chen Y, Ulrich JD, Randolph GJ, Zhang R, Fitzpatrick JAJ, Cashikar AG, Holtzman DM

EMDB-40711: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the chimeric Du-D1-Mo-D2 MXRA8 receptor
Method: single particle / : Zimmerman MI, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID)

PDB-8sqn: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the chimeric Du-D1-Mo-D2 MXRA8 receptor
Method: single particle / : Zimmerman MI, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID)

EMDB-28829: 
ELIC with Propylamine in SMA nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL

EMDB-28830: 
ELIC with Propylamine in saposin nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL

EMDB-28831: 
ELIC with Propylamine in spMSP1D1 nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL

EMDB-28832: 
Apo ELIC in spMSP1D1 nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL

PDB-8f32: 
ELIC with Propylamine in SMA nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL

PDB-8f33: 
ELIC with Propylamine in saposin nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL

PDB-8f34: 
ELIC with Propylamine in spMSP1D1 nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL

PDB-8f35: 
Apo ELIC in spMSP1D1 nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL

EMDB-41075: 
SARS-CoV-2 spike in complex with Fab 71281-33
Method: single particle / : Binshtein E, Crowe JE

EMDB-41076: 
SARS-CoV-2 spike in complex with Fab 71281-33 (2)
Method: single particle / : Binshtein E, Crowe JE

EMDB-28793: 
Voltage-gated potassium channel Kv3.1 with novel positive modulator (9M)-9-{5-chloro-6-[(3,3-dimethyl-2,3-dihydro-1-benzofuran-4-yl)oxy]-4-methylpyridin-3-yl}-2-methyl-7,9-dihydro-8H-purin-8-one (compound 4)
Method: single particle / : Chen Y, Ishchenko A

EMDB-28795: 
Voltage-gated potassium channel Kv3.1 apo
Method: single particle / : Chen Y, Ishchenko A

PDB-8f1c: 
Voltage-gated potassium channel Kv3.1 with novel positive modulator (9M)-9-{5-chloro-6-[(3,3-dimethyl-2,3-dihydro-1-benzofuran-4-yl)oxy]-4-methylpyridin-3-yl}-2-methyl-7,9-dihydro-8H-purin-8-one (compound 4)
Method: single particle / : Chen Y, Ishchenko A

PDB-8f1d: 
Voltage-gated potassium channel Kv3.1 apo
Method: single particle / : Chen Y, Ishchenko A

EMDB-29530: 
SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-29531: 
SARS-CoV-2 BQ.1.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-40240: 
SARS-CoV-2 BN.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

PDB-8fxb: 
SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

PDB-8fxc: 
SARS-CoV-2 BQ.1.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

PDB-8s9g: 
SARS-CoV-2 BN.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-16187: 
Wild tye immature Gag Structure
Method: subtomogram averaging / : Zhu Y, Chen L, Hardenbrook N, Zhang P

EMDB-16190: 
KAKA mutation immature Gag structure
Method: subtomogram averaging / : Zhu Y, Chen L, Hardenbrook N, Zhang P

EMDB-35326: 
Cryo-EM reconstruction of PBCV-1 capsid block 1
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35490: 
Cryo-EM reconstruction of PBCV-1 capsid block 2
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35585: 
Cryo-EM reconstruction of PBCV-1 capsid block 3
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35586: 
Cryo-EM reconstruction of PBCV-1 capsid block 4
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35589: 
Cryo-EM reconstruction of PBCV-1 capsid block 5
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35593: 
Cryo-EM reconstruction of PBCV-1 capsid block 6
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35594: 
Cryo-EM reconstruction of PBCV-1 capsid block 8
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35644: 
Cryo-EM reconstruction of PBCV-1 capsid block 9
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35645: 
Cryo-EM reconstruction of PBCV-1 capsid block 7
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35646: 
Cryo-EM reconstruction of PBCV-1 capsid block 10
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35647: 
Cryo-EM reconstruction of PBCV-1 capsid block 11
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35648: 
Cryo-EM reconstruction of PBCV-1 capsid block 12
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
