-Search query
-Search result
Showing all 46 items for (author: imada & k)

EMDB-35041: 
Cryo-EM structure of the J-K-St region of EMCV IRES in complex with eIF4G-HEAT1 and eIF4A
Method: single particle / : Suzuki H, Fujiyoshi Y, Imai S, Shimada I

EMDB-36046: 
Cryo-EM structure of the J-K-St region of EMCV IRES in complex with eIF4G-HEAT1 and eIF4A (J-K-St/eIF4G focused)
Method: single particle / : Suzuki H, Fujiyoshi Y, Imai S, Shimada I

PDB-8huj: 
Cryo-EM structure of the J-K-St region of EMCV IRES in complex with eIF4G-HEAT1 and eIF4A
Method: single particle / : Suzuki H, Fujiyoshi Y, Imai S, Shimada I

PDB-8j7r: 
Cryo-EM structure of the J-K-St region of EMCV IRES in complex with eIF4G-HEAT1 and eIF4A (J-K-St/eIF4G focused)
Method: single particle / : Suzuki H, Fujiyoshi Y, Imai S, Shimada I

EMDB-35399: 
Structure of human alpha-2/delta-1 with mirogabalin
Method: single particle / : Kozai D, Numoto N, Fujiyoshi Y

EMDB-35400: 
Structure of human alpha-2/delta-1 without mirogabalin
Method: single particle / : Kozai D, Numoto N, Fujiyoshi Y

PDB-8if3: 
Structure of human alpha-2/delta-1 with mirogabalin
Method: single particle / : Kozai D, Numoto N, Fujiyoshi Y

PDB-8if4: 
Structure of human alpha-2/delta-1 without mirogabalin
Method: single particle / : Kozai D, Numoto N, Fujiyoshi Y

EMDB-34203: 
CryoEM structure of pentameric MotA from Aquifex aeolicus
Method: single particle / : Nishikino T, Takekawa N, Kishikawa J, Hirose M, Onoe S, Kato T, Imada K

PDB-8gqy: 
CryoEM structure of pentameric MotA from Aquifex aeolicus
Method: single particle / : Nishikino T, Takekawa N, Kishikawa J, Hirose M, Onoe S, Kato T, Imada K

EMDB-30360: 
34-fold symmetry salmonella MS ring
Method: single particle / : Kawamoto A, Miyata T, Makino F, Kinoshita M, Minamino T, Imada K, Kato T, Namba K

EMDB-30361: 
11 fold-syymetry of salmonella MS ring
Method: single particle / : Kawamoto A, Miyata T, Makino F, Kinoshita M, Minamino T, Imada K, Kato T, Namba K

EMDB-30363: 
Without imposing symmetry of salmonella MS ring
Method: single particle / : Kawamoto A, Miyata T, Makino F, Kinoshita M, Minamino T, Imada K, Kato T, Namba K

EMDB-30613: 
Structure of the bacterial flagellar basal body
Method: single particle / : Kawamoto A, Miyata T, Makino F, Kinoshita M, Minamino T, Imada K, Kato T, Namba K

EMDB-30940: 
Salmonella flagella MS-ring protein FliF 1-503 having C33 symmetry
Method: single particle / : Makino F, Kawamoto A, Namba K

EMDB-30941: 
Salmonella flagella MS-ring protein FliF 1-456 having C35 symmetry
Method: single particle / : Makino F, Kawamoto A, Namba K

EMDB-30942: 
Salmonella flagella MS-ring protein FliF 1-456 having C34 symmetry
Method: single particle / : Makino F, Kawamoto A, Namba K
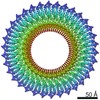
EMDB-30612: 
34-fold symmetry Salmonella S ring formed by full-length FliF
Method: single particle / : Kawamoto A, Miyata T

PDB-7d84: 
34-fold symmetry Salmonella S ring formed by full-length FliF
Method: single particle / : Kawamoto A, Miyata T, Makino F, Kinoshita M, Minamino T, Imada K, Kato T, Namba K

EMDB-30378: 
Salmonella FliF-FliG ring complex
Method: single particle / : Kawamoto A, Kato T, Kinoshita M, Minamino T, Namba K

EMDB-30379: 
11-fold symmetry of Salmonella FliF-FliG ring complex
Method: single particle / : Kawamoto A, Kato T, Kinoshita M, Minamino T, Namba K

EMDB-21027: 
Vibrio flagellar motor structure
Method: subtomogram averaging / : Zhu S, Nishikino T, Takekawa N, Terashima H, Kojima S, Imada K, Homma M, Liu J

EMDB-0919: 
Cryo-EM structure of the killifish CALHM1
Method: single particle / : Demura K, Kusakizako T, Shihoya W, Hiraizumi M, Shimada H, Yamashita K, Nishizawa T, Nureki O

EMDB-0920: 
Cryo-EM structure of the human CALHM2
Method: single particle / : Demura K, Kusakizako T, Shihoya W, Hiraizumi M, Shimada H, Yamashita K, Nishizawa T, Nureki O

EMDB-0921: 
Cryo-EM structure of the C. elegans CLHM-1
Method: single particle / : Demura K, Kusakizako T, Shihoya W, Hiraizumi M, Shimada H, Yamashita K, Nishizawa T, Nureki O

EMDB-0922: 
Cryo-EM structure of the CALHM chimeric construct (8-mer)
Method: single particle / : Demura K, Kusakizako T, Shihoya W, Hiraizumi M, Shimada H, Yamashita K, Nishizawa T, Nureki O

EMDB-0923: 
Cryo-EM structure of the CALHM chimeric construct (9-mer)
Method: single particle / : Demura K, Kusakizako T, Shihoya W, Hiraizumi M, Shimada H, Yamashita K, Nishizawa T, Nureki O

PDB-6lmt: 
Cryo-EM structure of the killifish CALHM1
Method: single particle / : Demura K, Kusakizako T, Shihoya W, Hiraizumi M, Shimada H, Yamashita K, Nishizawa T, Nureki O

PDB-6lmu: 
Cryo-EM structure of the human CALHM2
Method: single particle / : Demura K, Kusakizako T, Shihoya W, Hiraizumi M, Shimada H, Yamashita K, Nishizawa T, Nureki O

PDB-6lmv: 
Cryo-EM structure of the C. elegans CLHM-1
Method: single particle / : Demura K, Kusakizako T, Shihoya W, Hiraizumi M, Shimada H, Yamashita K, Nishizawa T, Nureki O

PDB-6lmw: 
Cryo-EM structure of the CALHM chimeric construct (8-mer)
Method: single particle / : Demura K, Kusakizako T, Shihoya W, Hiraizumi M, Shimada H, Yamashita K, Nishizawa T, Nureki O

PDB-6lmx: 
Cryo-EM structure of the CALHM chimeric construct (9-mer)
Method: single particle / : Demura K, Kusakizako T, Shihoya W, Hiraizumi M, Shimada H, Yamashita K, Nishizawa T, Nureki O

EMDB-0882: 
cryo-EM structure of TRPV3 in lipid nanodisc
Method: single particle / : Shimada H, Kusakizako T

PDB-6lgp: 
cryo-EM structure of TRPV3 in lipid nanodisc
Method: single particle / : Shimada H, Kusakizako T, Nishizawa T, Nureki O

PDB-6kmf: 
FimA type V pilus from P.gingivalis
Method: single particle / : Shibata S, Shoji M, Matsunami H, Matthews M, Imada K, Nakayama K, Wolf M

EMDB-9996: 
Complex of yeast cytoplasmic dynein MTBD-High and MT without DTT
Method: helical / : Komori Y, Nishida N, Shimada I, Kikkawa M

EMDB-9997: 
Complex of yeast cytoplasmic dynein MTBD-High and MT with DTT
Method: helical / : Komori Y, Nishida N

PDB-6kio: 
Complex of yeast cytoplasmic dynein MTBD-High and MT without DTT
Method: helical / : Komori Y, Nishida N, Shimada I, Kikkawa M

PDB-6kiq: 
Complex of yeast cytoplasmic dynein MTBD-High and MT with DTT
Method: helical / : Komori Y, Nishida N, Shimada I, Kikkawa M

PDB-6jzr: 
Structure of the bacterial flagellar polyrod
Method: helical / : Saijo-Hamano Y, Matsunami H, Namba K, Imada K

PDB-6jzt: 
Structure of the bacterial flagellar hook from Salmonella typhimurium
Method: helical / : Saijo-Hamano Y, Matsunami H, Namba K, Imada K

EMDB-3098: 
Cryo-EM structure of bovine FoF1 ATP synthase
Method: single particle / : Hauer F, Gerle C, Fischer N, Oshima A, Shinzawa-Itoh K, Shimada S, Yokoyama K, Fujiyoshi Y, Stark H

EMDB-1132: 
A partial atomic structure for the flagellar hook of Salmonella typhimurium.
Method: helical / : Shaikh TR, Thomas DR, Chen JZ, Samatey FA, Matsunami H, Imada K, Namba K, DeRosier DJ

PDB-2bgy: 
Fit of the x-ray structure of the baterial flagellar hook fragment flge31 into an EM map from the hook of Caulobacter crescentus.
Method: electron crystallography / : Shaikh TR, Thomas DR, Chen JZ, Samatey FA, Matsunami H, Imada K, Namba K, DeRosier DJ

PDB-2bgz: 
ATOMIC MODEL OF THE BACTERIAL FLAGELLAR BASED ON DOCKING AN X-RAY DERIVED HOOK STRUCTURE INTO AN EM MAP.
Method: electron crystallography / : Shaikh TR, Thomas DR, Chen JZ, Samatey FA, Matsunami H, Imada K, Namba K, Derosier DJ
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
