-Search query
-Search result
Showing 1 - 50 of 2,836 items for (author: hui & w)

EMDB entry, No image
EMDB-37362: 
CryoEM structure of human PI3K-alpha (P85/P110-H1047R) with QR-7909 binding at an allosteric site
Method: single particle / : Huang X, Ren X, Zhong W

EMDB entry, No image
EMDB-37363: 
CryoEM structure of human PI3K-alpha (P85/P110-H1047R) with QR-8557 binding at an allosteric site
Method: single particle / : Huang X, Ren X, Zhong W

PDB-8w9a: 
CryoEM structure of human PI3K-alpha (P85/P110-H1047R) with QR-7909 binding at an allosteric site
Method: single particle / : Huang X, Ren X, Zhong W

PDB-8w9b: 
CryoEM structure of human PI3K-alpha (P85/P110-H1047R) with QR-8557 binding at an allosteric site
Method: single particle / : Huang X, Ren X, Zhong W

EMDB entry, No image
EMDB-37529: 
Structure of DDM1-nucleosome complex in the apo state
Method: single particle / : Liu Y, Zhang Z, Du J

EMDB entry, No image
EMDB-37533: 
Structure of DDM1-nucleosome complex in ADP state
Method: single particle / : Liu Y, Zhang Z, Du J

EMDB entry, No image
EMDB-37535: 
Structure of DDM1-nucleosome complex in ADP-BeFx state
Method: single particle / : Liu Y, Zhang Z, Du J

EMDB entry, No image
EMDB-37537: 
Structure of DDM1-nucleosome complex in the ADP-BeFx state with DDM1 bound to SHL2 and SHL-2
Method: single particle / : Liu Y, Zhang Z, Du J

EMDB entry, No image
EMDB-37538: 
Structure of nucleosome core particle of Arabidopsis thaliana
Method: single particle / : Liu Y, Zhang Z, Du J

PDB-8wh5: 
Structure of DDM1-nucleosome complex in the apo state
Method: single particle / : Liu Y, Zhang Z, Du J

PDB-8wh8: 
Structure of DDM1-nucleosome complex in ADP state
Method: single particle / : Liu Y, Zhang Z, Du J

PDB-8wh9: 
Structure of DDM1-nucleosome complex in ADP-BeFx state
Method: single particle / : Liu Y, Zhang Z, Du J

PDB-8wha: 
Structure of DDM1-nucleosome complex in the ADP-BeFx state with DDM1 bound to SHL2 and SHL-2
Method: single particle / : Liu Y, Zhang Z, Du J

PDB-8whb: 
Structure of nucleosome core particle of Arabidopsis thaliana
Method: single particle / : Liu Y, Zhang Z, Du J

EMDB entry, No image
EMDB-37240: 
SARS-CoV-2 Omicron spike in complex with 5817 Fab
Method: single particle / : Cao L, Wang X

EMDB entry, No image
EMDB-37241: 
The interface structure of Omicron RBD binding to 5817 Fab
Method: single particle / : Cao L, Wang X

PDB-8khc: 
SARS-CoV-2 Omicron spike in complex with 5817 Fab
Method: single particle / : Cao L, Wang X

PDB-8khd: 
The interface structure of Omicron RBD binding to 5817 Fab
Method: single particle / : Cao L, Wang X

EMDB-36918: 
Cryo-EM structure of Oryza sativa HKT2;1 at 2.5 angstrom
Method: single particle / : Wang X, Shen X, Qu Y, Wang C, Shen H

EMDB-36919: 
Cryo-EM structure of Oryza sativa HKT2;2/1 at 2.3 angstrom
Method: single particle / : Wang X, Shen X, Qu Y, Wang C, Shen H

PDB-8k66: 
Cryo-EM structure of Oryza sativa HKT2;1 at 2.5 angstrom
Method: single particle / : Wang X, Shen X, Qu Y, Wang C, Shen H

PDB-8k69: 
Cryo-EM structure of Oryza sativa HKT2;2/1 at 2.3 angstrom
Method: single particle / : Wang X, Shen X, Qu Y, Wang C, Shen H

EMDB-35906: 
cryo-EM structure of human EMC
Method: single particle / : Li M, Zhang C, Wu J, Lei M
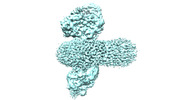
EMDB-35907: 
cryo-EM structure of human EMC and VDAC
Method: single particle / : Li M, Zhang C, Wu J, Lei M

PDB-8j0o: 
cryo-EM structure of human EMC and VDAC
Method: single particle / : Li M, Zhang C, Wu J, Lei M

EMDB-38497: 
Cryo-EM structure of the ClpP degradation system in Streptomyces hawaiiensis
Method: single particle / : Xu X, Long F

EMDB-38535: 
Cryo-EM structure of the ClpC1:ClpP1P2 degradation complex in Streptomyces hawaiiensis
Method: single particle / : Xu X, Long F

EMDB-38536: 
Cryo-EM structure of the ClpC1:ClpP1P2 degradation complex in Streptomyces hawaiiensis
Method: single particle / : Xu X, Long F

EMDB-38537: 
Cryo-EM structure of ClpP1P2 in complex with ADEP1 from Streptomyces hawaiiensis
Method: single particle / : Xu X, Long F

PDB-8xn4: 
Cryo-EM structure of the ClpP degradation system in Streptomyces hawaiiensis
Method: single particle / : Xu X, Long F

PDB-8xon: 
Cryo-EM structure of the ClpC1:ClpP1P2 degradation complex in Streptomyces hawaiiensis
Method: single particle / : Xu X, Long F

PDB-8xoo: 
Cryo-EM structure of the ClpC1:ClpP1P2 degradation complex in Streptomyces hawaiiensis
Method: single particle / : Xu X, Long F

PDB-8xop: 
Cryo-EM structure of ClpP1P2 in complex with ADEP1 from Streptomyces hawaiiensis
Method: single particle / : Xu X, Long F
EMDB-39012: 
Representative tomogram of primary glioblastoma stem cell with circular inter-mitochondrial junctions.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39015: 
Representative tomogram of microglia cell with nanotunnel-like structures resembling mitochondrial fission.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT
EMDB-39019: 
Representative tomogram of glioblastoma cell with nanotunnel-like structure and inter-mitochondrial junction.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39021: 
Representative tomogram of normal human astrocyte with nanotunnel-like structure which is an extension of the mitochondrial outer membrane.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39023: 
Representative tomogram of primary glioblastoma differentiated cell with parallel inter-mitochondrial junction.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39024: 
Representative tomogram of primary glioblastoma stem cell with clustered mitochondria bearing various long-short axis ratios.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-33347: 
Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Postfusion state)
Method: single particle / : Wu Z, Yu Z, Tan S, Lu J, Lu G, Lin J

PDB-7xog: 
Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Postfusion state)
Method: single particle / : Wu Z, Yu Z, Tan S, Lu J, Lu G, Lin J

EMDB-35449: 
Eaf3 CHD domain bound to the nucleosome
Method: single particle / : Zhang Y, Gang C

EMDB-35450: 
Cryo-EM structure of the Rpd3S core complex
Method: single particle / : Zhang Y, Gang C

EMDB-35455: 
Rpd3S bound to the nucleosome
Method: single particle / : Zhang Y, Gang C

EMDB-37386: 
The cryo-EM structure of the Nicotiana tabacum PEP-PAP
Method: single particle / : Wu XX, Zhang Y

EMDB-37387: 
The cryo-EM structure of the Nicotiana tabacum PEP-PAP-TEC1
Method: single particle / : Wu XX, Zhang Y
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search







 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
