-Search query
-Search result
Showing 1 - 50 of 141 items for (author: homa & fl)

EMDB-19250: 
Pseudoatomic model of a second-order Sierpinski triangle formed by the citrate synthase from Synechococcus elongatus
Method: single particle / : Lo YK, Bohn S, Sendker FL, Schuller JM, Hochberg G

EMDB-19251: 
Structure of a first order Sierpinski triangle formed by the H369R mutant of the citrate synthase from Synechococcus elongatus
Method: single particle / : Lo YK, Bohn S, Sendker FL, Schuller JM, Hochberg G

EMDB-16805: 
Cryo-EM structure of PcrV/Fab(30-B8)
Method: single particle / : Yuan B, Simonis A, Marlovits TC

EMDB-16807: 
Cryo-EM structure of PcrV/Fab(11-E5)
Method: single particle / : Yuan B, Simonis A, Marlovits TC

PDB-8cr9: 
Cryo-EM structure of PcrV/Fab(30-B8)
Method: single particle / : Yuan B, Simonis A, Marlovits TC

PDB-8crb: 
Cryo-EM structure of PcrV/Fab(11-E5)
Method: single particle / : Yuan B, Simonis A, Marlovits TC

EMDB-17154: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (consensus and constituent map 1)
Method: single particle / : Stoos L, Michael AK, Kempf G, Cavadini S, Thoma NH

EMDB-17155: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL-6.2 (DNA conformation 1)
Method: single particle / : Michael AK, Stoos L, Kempf G, Cavadini S, Thoma NH

EMDB-17156: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL-6.2 (DNA conformation 2)
Method: single particle / : Michael AK, Stoos L, Kempf G, Cavadini S, Thoma NH

EMDB-17157: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (composite map)
Method: single particle / : Stoos L, Michael AK, Kempf G, Cavadini S, Thoma NH

EMDB-17158: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (constituent map 2 from additional focus classification on PAS domains)
Method: single particle / : Stoos L, Michael AK, Kempf G, Cavadini S, Thoma NH

EMDB-17159: 
Cryo-EM map of MYC-MAX-OCT4-LIN28 complex
Method: single particle / : Michael AK, Kempf G, Cavadini S, Thoma NH

EMDB-17160: 
Cryo-EM structure of CLOCK-BMAL1 bound to the native Por enhancer nucleosome (map 2, additional 3D classification and flexible refinement)
Method: single particle / : Michael AK, Stoos L, Kempf G, Cavadini S, Thoma N

EMDB-17161: 
Cryo-EM structure of CLOCK-BMAL1 bound to the native Por enhancer nucleosome (map 1)
Method: single particle / : Michael AK, Stoos L, Cavadini S, Kempf G

EMDB-17162: 
MAX-MAX bound to a nucleosome at SHL+5.1 and SHL-6.9.
Method: single particle / : Stoos L, Kempf G, Kater L, Thoma NH

EMDB-17183: 
OCT4 and MYC-MAX co-bound to a nucleosome
Method: single particle / : Michael AK, Stoos L, Kempf G, Cavadini S, Thoma N

EMDB-17184: 
MYC-MAX bound to a nucleosome at SHL+5.8
Method: single particle / : Stoos L, Michael AK, Kempf G, Kater L, Cavadini S, Thoma N

PDB-8osj: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL-6.2 (DNA conformation 1)
Method: single particle / : Michael AK, Stoos L, Kempf G, Cavadini S, Thoma NH

PDB-8osk: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (composite map)
Method: single particle / : Stoos L, Michael AK, Kempf G, Cavadini S, Thoma NH

PDB-8osl: 
Cryo-EM structure of CLOCK-BMAL1 bound to the native Por enhancer nucleosome (map 2, additional 3D classification and flexible refinement)
Method: single particle / : Michael AK, Stoos L, Kempf G, Cavadini S, Thoma N

PDB-8ots: 
OCT4 and MYC-MAX co-bound to a nucleosome
Method: single particle / : Michael AK, Stoos L, Kempf G, Cavadini S, Thoma N

PDB-8ott: 
MYC-MAX bound to a nucleosome at SHL+5.8
Method: single particle / : Stoos L, Michael AK, Kempf G, Kater L, Cavadini S, Thoma N

EMDB-15900: 
Subtomogram average of the respiratory supercomplex from Tetrahymena thermophila mitochondria
Method: subtomogram averaging / : Muhleip A, Amunts A

EMDB-15865: 
Cryo-EM structure of NADH:ubiquinone oxidoreductase (complex-I) from respiratory supercomplex of Tetrahymena thermophila
Method: single particle / : Muhleip A, Kock Flygaard R, Amunts A

EMDB-15866: 
Cryo-EM structure of succinate dehydrogenase complex (complex-II) in respiratory supercomplex of Tetrahymena thermophila
Method: single particle / : Muhleip A, Kock Flygaard R, Baradaran R, Amunts A

EMDB-15867: 
Cryo-EM structure of cytochrome c oxidase dimer (complex IV) from respiratory supercomplex of Tetrahymena thermophila
Method: single particle / : Muhleip A, Kock Flygaard R, Amunts A

EMDB-15868: 
Cryo-EM structure of cytochrome bc1 complex (complex-III) from respiratory supercomplex of Tetrahymena thermophila
Method: single particle / : Muhleip A, Kock Flygaard R, Amunts A

PDB-8b6f: 
Cryo-EM structure of NADH:ubiquinone oxidoreductase (complex-I) from respiratory supercomplex of Tetrahymena thermophila
Method: single particle / : Muhleip A, Kock Flygaard R, Amunts A

PDB-8b6g: 
Cryo-EM structure of succinate dehydrogenase complex (complex-II) in respiratory supercomplex of Tetrahymena thermophila
Method: single particle / : Muhleip A, Kock Flygaard R, Baradaran R, Amunts A

PDB-8b6h: 
Cryo-EM structure of cytochrome c oxidase dimer (complex IV) from respiratory supercomplex of Tetrahymena thermophila
Method: single particle / : Muhleip A, Kock Flygaard R, Amunts A

PDB-8b6j: 
Cryo-EM structure of cytochrome bc1 complex (complex-III) from respiratory supercomplex of Tetrahymena thermophila
Method: single particle / : Muhleip A, Kock Flygaard R, Amunts A

EMDB-16687: 
Carin1 bacteriophage mature capsid
Method: single particle / : d'Acapito A, Neumann E, Schoehn G

EMDB-16688: 
Carin1 bacteriophage portal assembly
Method: single particle / : d'Acapito A, Neumann E, Schoehn G

EMDB-16689: 
Carin 1 bacteriophage tail, connector and tail fibers assembly
Method: single particle / : d'Acapito A, Neumann E, Schoehn G

PDB-8cjz: 
Carin1 bacteriophage mature capsid
Method: single particle / : d'Acapito A, Neumann E, Schoehn G

PDB-8ck0: 
Carin1 bacteriophage portal assembly
Method: single particle / : d'Acapito A, Neumann E, Schoehn G

PDB-8ck1: 
Carin 1 bacteriophage tail, connector and tail fibers assembly
Method: single particle / : d'Acapito A, Neumann E, Schoehn G
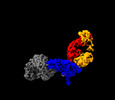
EMDB-28523: 
Structure of interleukin receptor common gamma chain (IL2Rgamma) in complex with two antibodies
Method: single particle / : Franklin MC, Romero Hernandez A

PDB-8epa: 
Structure of interleukin receptor common gamma chain (IL2Rgamma) in complex with two antibodies
Method: single particle / : Franklin MC, Romero Hernandez A

EMDB-26604: 
H1 Solomon Islands 2006 hemagglutinin in complex with Ab111
Method: single particle / : Windsor IW, Caradonna TM, Schmidt AG
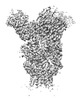
EMDB-26605: 
H1 Solomon Islands 2006 hemagglutinin in complex with Ab109
Method: single particle / : Windsor IW, Caradonna TM, Schmidt AG

PDB-7umm: 
H1 Solomon Islands 2006 hemagglutinin in complex with Ab109
Method: single particle / : Windsor IW, Caradonna TM, Schmidt AG

EMDB-13608: 
Focused reconstruction of Haliangium ochraceum encapsulated ferritin cargo within the encapsulin nano compartment
Method: single particle / : Marles-Wright J, Basle A, Ross J, Clarke DJ

EMDB-24783: 
CryoEM structure of Vibrio cholerae transposon Tn6677 AAA+ ATPase TnsC
Method: single particle / : Hoffmann FT, Kim M, Beh LY, Wang J, Vo PLH, Gelsinger DR, Acree C, Mohabir JT, Fernandez IS, Sternberg SH

EMDB-26476: 
VchTnsC AAA+ ATPase with DNA, single heptamer
Method: single particle / : Fernandez IS, Sternberg SH

EMDB-26477: 
VchTnsC AAA+ with DNA (double heptamer)
Method: single particle / : Fernandez IS, Sternberg SH

PDB-7rzy: 
CryoEM structure of Vibrio cholerae transposon Tn6677 AAA+ ATPase TnsC
Method: single particle / : Hoffmann FT, Kim M, Beh LY, Wang J, Vo PLH, Gelsinger DR, Acree C, Mohabir JT, Fernandez IS, Sternberg SH

PDB-7ufi: 
VchTnsC AAA+ ATPase with DNA, single heptamer
Method: single particle / : Fernandez IS, Sternberg SH

PDB-7ufm: 
VchTnsC AAA+ with DNA (double heptamer)
Method: single particle / : Fernandez IS, Sternberg SH

EMDB-12853: 
Icosahedral cryo-EM reconstruction of Haliangium ochraceum encapsulin
Method: single particle / : Marles-Wright J, Basle A, Clarke DJ, Ross J
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
