-Search query
-Search result
Showing all 32 items for (author: hess & s)

EMDB-29783: 
Cryo-EM structure of T/F100 SOSIP.664 HIV-1 Env trimer with LMHS mutations in complex with 8ANC195 and 10-1074
Method: single particle / : Chen Y, Zhou F, Huang R, Tolbert W, Pazgier M

EMDB-41613: 
Cryo-EM structure of BG505 SOSIP.664 HIV-1 Env trimer in complex with temsavir, 8ANC195, and 10-1074
Method: single particle / : Tolbert WD, Pozharski E, Pazgier M

PDB-8g6u: 
Cryo-EM structure of T/F100 SOSIP.664 HIV-1 Env trimer with LMHS mutations in complex with 8ANC195 and 10-1074
Method: single particle / : Chen Y, Zhou F, Huang R, Tolbert W, Pazgier M

PDB-8ttw: 
Cryo-EM structure of BG505 SOSIP.664 HIV-1 Env trimer in complex with temsavir, 8ANC195, and 10-1074
Method: single particle / : Tolbert WD, Pozharski E, Pazgier M

EMDB-28378: 
Structure of the C3bB proconvertase in complex with lufaxin and factor Xa
Method: single particle / : Andersen JF, Lei H

PDB-8eok: 
Structure of the C3bB proconvertase in complex with lufaxin and factor Xa
Method: single particle / : Andersen JF, Lei H

EMDB-28279: 
Structure of the C3bB proconvertase in complex with lufaxin
Method: single particle / : Andersen JF, Lei H

PDB-8enu: 
Structure of the C3bB proconvertase in complex with lufaxin
Method: single particle / : Andersen JF, Lei H

EMDB-27596: 
Cryo-EM structure of T/F100 SOSIP.664 HIV-1 Env trimer in complex with 8ANC195 and 10-1074
Method: single particle / : Chen Y, Zhou F, Huang R, Tolbert W, Pazgier M

PDB-8dok: 
Cryo-EM structure of T/F100 SOSIP.664 HIV-1 Env trimer in complex with 8ANC195 and 10-1074
Method: single particle / : Chen Y, Zhou F, Huang R, Tolbert W, Pazgier M

EMDB-27103: 
Cryo-EM structure of T/F100 SOSIP.664 HIV-1 Env trimer with LMHS mutations in complex with Temsavir, 8ANC195, and 10-1074
Method: single particle / : Chen Y, Pozharski E, Tolbert W, Pazgier M

PDB-8czz: 
Cryo-EM structure of T/F100 SOSIP.664 HIV-1 Env trimer with LMHS mutations in complex with Temsavir, 8ANC195, and 10-1074
Method: single particle / : Chen Y, Pozharski E, Tolbert W, Pazgier M
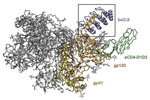
EMDB-26157: 
Cryo-EM structure of BG505 SOSIP HIV-1 Env trimer in complex with CD4 receptor (D1D2) and broadly neutralizing darpin bnD.9
Method: single particle / : Cerutti G, Gorman J

PDB-7txd: 
Cryo-EM structure of BG505 SOSIP HIV-1 Env trimer in complex with CD4 receptor (D1D2) and broadly neutralizing darpin bnD.9
Method: single particle / : Cerutti G, Gorman J, Kwong PD, Shapiro L

EMDB-33233: 
Cryo-EM structure of EDS1 and SAG101 with ATP-APDR
Method: single particle / : Huang SJ, Jia AL, Han ZF, Chai JJ

PDB-7xjp: 
Cryo-EM structure of EDS1 and SAG101 with ATP-APDR
Method: single particle / : Huang SJ, Jia AL, Han ZF, Chai JJ

EMDB-33144: 
Cryo-EM structure of EDS1 and PAD4
Method: single particle / : Huang SJ, Jia AL, Sun Y, Han ZF, Chai JJ

PDB-7xdd: 
Cryo-EM structure of EDS1 and PAD4
Method: single particle / : Huang SJ, Jia AL, Sun Y, Han ZF, Chai JJ

EMDB-12700: 
Structure of the C9orf72-SMCR8 complex
Method: single particle / : Noerpel J, Cavadini S, Schenk AD, Graff-Meyer A, Chao J, Bhaskar V

PDB-7o2w: 
Structure of the C9orf72-SMCR8 complex
Method: single particle / : Noerpel J, Cavadini S, Schenk AD, Graff-Meyer A, Chao J, Bhaskar V

EMDB-10049: 
THE STRUCTURE OF BD OXIDASE FROM ESCHERICHIA COLI
Method: single particle / : Rasmussen T, Boettcher B, Thesseling A, Friedrich T

PDB-6rx4: 
THE STRUCTURE OF BD OXIDASE FROM ESCHERICHIA COLI
Method: single particle / : Rasmussen T, Boettcher B, Thesseling A, Friedrich T

EMDB-4584: 
Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel J, Wollweber F, Pfitzner A, Muehleip A, Sanchez R, Kudryashev M, Chiaruttin N, Lilie H, Schleger J, Rosenbaum E, Hessenberger M, Matthaeus C, Noe F, Roux A, van der Laan M, Kuehlbrandt W, Daumke O
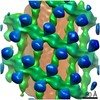
EMDB-10062: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuehlbrandt W, Daumke O
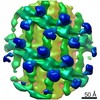
EMDB-10063: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPgammaS bound state
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

EMDB-10064: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

EMDB-10065: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes in the GTPgammaS bound state
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

PDB-6rzt: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuehlbrandt W, Daumke O

PDB-6rzu: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPgammaS bound state
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

PDB-6rzv: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

PDB-6rzw: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes in the GTPgammaS bound state
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

EMDB-3401: 
Electron cryo-microscopy of CSN-SCF-N8 complex
Method: single particle / : Mosadeghi R, Reichermeier KM, Winkler M, Schreiber A, Reitsma JM, Zhang Y, Stengel F, Cao J, Kim M, Sweredoski MJ, Hess S, Leitner A, Aebersold R, Peter M, Deshaies RJ, Enchev RI
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
