-Search query
-Search result
Showing 1 - 50 of 296 items for (author: henderson & r)

EMDB-40853: 
CH505 Disulfide Stapled SOSIP Bound to b12 Fab
Method: single particle / : Henderson R
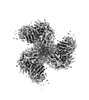
EMDB-40854: 
CH505 Disulfide Stapled SOSIP Bound to CH235.12 Fab
Method: single particle / : Henderson R

PDB-8sxi: 
CH505 Disulfide Stapled SOSIP Bound to b12 Fab
Method: single particle / : Henderson R

PDB-8sxj: 
CH505 Disulfide Stapled SOSIP Bound to CH235.12 Fab
Method: single particle / : Henderson R

EMDB-17958: 
Structure of DPS determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

EMDB-17959: 
Structure of bacterial ribosome determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

EMDB-17960: 
Structure of GABAAR determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

EMDB-17961: 
Structure of mouse heavy-chain apoferritin determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

EMDB-17962: 
Structure of catalase determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

EMDB-17963: 
Structure of AHIR determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

EMDB-17964: 
Structure of GAPDH determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

EMDB-17965: 
Structure of E. coli glutamine synthetase determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

EMDB-17966: 
Structure of human apo ALDH1A1 determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

EMDB-17967: 
Structure of PaaZ determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

EMDB-17968: 
Structure of lumazine synthase determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

PDB-8pv9: 
Structure of DPS determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

PDB-8pva: 
Structure of bacterial ribosome determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

PDB-8pvb: 
Structure of GABAAR determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

PDB-8pvc: 
Structure of mouse heavy-chain apoferritin determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

PDB-8pvd: 
Structure of catalase determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

PDB-8pve: 
Structure of AHIR determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

PDB-8pvf: 
Structure of GAPDH determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

PDB-8pvg: 
Structure of E. coli glutamine synthetase determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

PDB-8pvh: 
Structure of human apo ALDH1A1 determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

PDB-8pvi: 
Structure of PaaZ determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

PDB-8pvj: 
Structure of lumazine synthase determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

EMDB-27703: 
Structure of RBD directed antibody DH1047 in complex with SARS-CoV-2 spike: Local refinement of RBD-Fab interace
Method: single particle / : May AJ, Manne K, Acharya P

PDB-8dtk: 
Structure of RBD directed antibody DH1047 in complex with SARS-CoV-2 spike: Local refinement of RBD-Fab interace
Method: single particle / : May AJ, Manne K, Acharya P

PDB-7tcn: 
Cryo-EM structure of CH235.12 in complex with HIV-1 Env trimer CH505TF.N279K.SOSIP.664 with high-mannose glycans
Method: single particle / : Manne K, Henderson R, Acharya P

PDB-7tco: 
Cryo-EM structure of CH235.12 in complex with HIV-1 Env trimer CH505TF.N279K.G458Y.SOSIP.664 with high-mannose glycans
Method: single particle / : Manne K, Henderson R, Acharya P
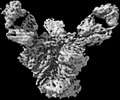
EMDB-27624: 
Cryo-EM structure of HIV-1 Env(CH848 10.17 DS.SOSIP_DT) in complex with DH1030.1 Fab
Method: single particle / : Gobeil S, Acharya P
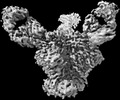
EMDB-27628: 
Cryo-EM structure of HIV-1 Env(BG505.T332N SOSIP) in complex with DH1030.1 Fab
Method: single particle / : Gobeil S, Acharya P

EMDB-40273: 
CryoEM structure of VRC01-CH848.0358.80
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40274: 
CryoEM structure of VRC01-CH848.0836.10
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40275: 
CryoEM structure of DH270.6-CH848.0526.25
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40277: 
CryoEM structure of DH270.6-CH848.10.17
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40278: 
CryoEM structure of DH270.5-CH848.10.17
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40279: 
CryoEM structure of VRC01-CH848.10.17
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40280: 
CryoEM structure of DH270.4-CH848.10.17
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40281: 
CryoEM structure of VRC01-CH848.0526.25
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40282: 
CryoEM structure of DH270.UCA.G57R-CH848.10.17DT
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40283: 
CryoEM structure of DH270.UCA-CH848.10.17DT
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40284: 
CryoEM structure of DH270.3-CH848.10.17
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40285: 
CryoEM structure of DH270.I5.6-CH848.10.17
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40286: 
CryoEM structure of DH270.I4.6-CH848.10.17
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40287: 
CryoEM structure of DH270.I3-CH848.10.17
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40288: 
CryoEM structure of DH270.I2-CH848.10.17
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40289: 
CryoEM structure of DH270.2-CH848.10.17
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40290: 
CryoEM structure of DH270.1-CH848.10.17
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P

EMDB-40291: 
CryoEM structure of DH270.I1.6-CH848.10.17
Method: single particle / : Henderson R, Zhou Y, Stalls V, Bartesaghi B, Acharya P
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
