-Search query
-Search result
Showing 1 - 50 of 133 items for (author: hassan & d)

EMDB-43320: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

EMDB-43321: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (conformation 2)
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

EMDB-43322: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (conformation 1)
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

EMDB-43323: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (focused refinement of RBD and mouse ACE2)
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

EMDB-43324: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with human ACE2
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

EMDB-43325: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with human ACE2 (focused refinement of RBD and ACE2)
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

EMDB-43326: 
Negative Stain EM Reconstructions of SARS-CoV-2 spike proteins mixed with polyclonal antibodies from donor 4.
Method: single particle / : Mannar D, Zhu X, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

PDB-8vkk: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

PDB-8vkl: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (conformation 2)
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

PDB-8vkm: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (conformation 1)
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

PDB-8vkn: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (focused refinement of RBD and mouse ACE2)
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

PDB-8vko: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with human ACE2
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

PDB-8vkp: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with human ACE2 (focused refinement of RBD and ACE2)
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

EMDB-40711: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the chimeric Du-D1-Mo-D2 MXRA8 receptor
Method: single particle / : Zimmerman MI, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID)

PDB-8sqn: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the chimeric Du-D1-Mo-D2 MXRA8 receptor
Method: single particle / : Zimmerman MI, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID)

EMDB-27272: 
CryoEM structure of Western equine encephalitis virus VLP
Method: single particle / : Zimmerman MI, Fremont DH
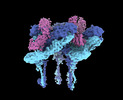
EMDB-27271: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the avian MXRA8 receptor
Method: single particle / : Zimmerman MI, Fremont DH
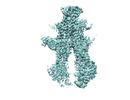
EMDB-41004: 
Heterodimeric ABC transporter BmrCD in the inward-facing conformation bound to ATP: BmrCD_IF-ATP2
Method: single particle / : Tang Q, Mchaourab H

PDB-8t3k: 
Heterodimeric ABC transporter BmrCD in the inward-facing conformation bound to ATP: BmrCD_IF-ATP2
Method: single particle / : Tang Q, Mchaourab H
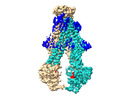
EMDB-29362: 
Heterodimeric ABC transporter BmrCD in the inward-facing conformation bound to ATP: BmrCD_IF-ATP
Method: single particle / : Qingyu T, Hassane M

PDB-8fpf: 
Heterodimeric ABC transporter BmrCD in the inward-facing conformation bound to ATP: BmrCD_IF-ATP
Method: single particle / : Qingyu T, Hassane M

EMDB-29087: 
Heterodimeric ABC transporter BmrCD in the occluded conformation bound to ATP: BmrCD_OC-ATP
Method: single particle / : Tang Q, Mchaourab H

EMDB-29297: 
Structure0915
Method: single particle / : Qingyu T, Mchaourab H

EMDB-40908: 
Heterodimeric ABC transporter BmrCD in the inward-facing conformation bound to substrate and ATP: BmrCD_IF-1HT/ATP
Method: single particle / : Tang Q, Mchaourab H

EMDB-40974: 
BmrCD_OC-ADPVi
Method: single particle / : Tang Q, Hassane M

EMDB-41058: 
Heterodimeric ABC transporter BmrCD in the inward-facing conformation bound to substrate and ADPVi: BmrCD_IF-HT/ADPVi
Method: single particle / : Qingyu T, Hassane M

PDB-8fhk: 
Heterodimeric ABC transporter BmrCD in the occluded conformation bound to ATP: BmrCD_OC-ATP
Method: single particle / : Tang Q, Mchaourab H

PDB-8fmv: 
Heterodimeric ABC transporter BmrCD in the inward-facing conformation bound to substrate and ATP: BmrCD_IF-2HT/ATP
Method: single particle / : Tang Q, Mchaourab H

PDB-8szc: 
Heterodimeric ABC transporter BmrCD in the inward-facing conformation bound to substrate and ATP: BmrCD_IF-1HT/ATP
Method: single particle / : Tang Q, Mchaourab H

PDB-8t1p: 
Heterodimeric ABC transporter BmrCD in the occluded conformation bound to ADPVi: BmrCD_OC-ADPVi
Method: single particle / : Tang Q, Mchaourab HS

EMDB-28644: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the avian MXRA8 receptor
Method: single particle / : Zimmerman MI, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID)

EMDB-13981: 
Human mitochondrial ribosome in complex with mRNA, A/A-, P/P- and E/E-tRNAs at 2.63 A resolution
Method: single particle / : Singh V, Itoh Y, Andrell J, Aibara S, Amunts A

PDB-7qi5: 
Human mitochondrial ribosome in complex with mRNA, A/A-, P/P- and E/E-tRNAs at 2.63 A resolution
Method: single particle / : Singh V, Itoh Y, Amunts A

EMDB-16268: 
E. coli BAM complex (BamABCDE) bound to darobactin B
Method: single particle / : Horne JE, Fenn KL, Radford SE, Ranson NA

EMDB-16282: 
E. coli BAM complex (BamABCDE) wild-type
Method: single particle / : Machin JM, Radford SE, Ranson NA

PDB-8bvq: 
E. coli BAM complex (BamABCDE) bound to darobactin B
Method: single particle / : Horne JE, Fenn KL, Radford SE, Ranson NA

PDB-8bwc: 
E. coli BAM complex (BamABCDE) wild-type
Method: single particle / : Machin JM, Radford SE, Ranson NA

EMDB-13982: 
Human mitochondrial ribosome in complex with mRNA, A/P- and P/E-tRNAs at 2.98 A resolution
Method: single particle / : Singh V, Itoh Y, Amunts A, Aibara S

PDB-7qi6: 
Human mitochondrial ribosome in complex with mRNA, A/P- and P/E-tRNAs at 2.98 A resolution
Method: single particle / : Singh V, Itoh Y, Amunts A

EMDB-29714: 
Cryo-EM structure of DDB1deltaB-DDA1-DCAF16-BRD4(BD2)-MMH2
Method: single particle / : Ma MW, Hunkeler M, Jin CY, Fischer ES

PDB-8g46: 
Cryo-EM structure of DDB1deltaB-DDA1-DCAF16-BRD4(BD2)-MMH2
Method: single particle / : Ma MW, Hunkeler M, Jin CY, Fischer ES

EMDB-27527: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with human ACE2
Method: single particle / : Zhu X, Saville JW, Mannar D, Berezuk AM, Cholak S, Tuttle KS, Vahdatihassani F, Subramaniam S

EMDB-27528: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with human ACE2 (focused refinement of RBD and ACE2)
Method: single particle / : Zhu X, Saville JW, Mannar D, Berezuk AM, Cholak S, Tuttle KS, Vahdatihassani F, Subramaniam S

EMDB-27529: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with mouse ACE2
Method: single particle / : Zhu X, Saville JW, Mannar D, Berezuk AM, Cholak S, Tuttle KS, Vahdatihassani F, Subramaniam S

EMDB-27530: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with mouse ACE2 (focused refinement of RBD and ACE2)
Method: single particle / : Zhu X, Saville JW, Mannar D, Berezuk AM, Cholak S, Tuttle KS, Vahdatihassani F, Subramaniam S

EMDB-27531: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.1 spike protein in complex with mouse ACE2
Method: single particle / : Zhu X, Saville JW, Mannar D, Berezuk AM, Cholak S, Tuttle KS, Vahdatihassani F, Subramaniam S

EMDB-27532: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.1 spike protein in complex with mouse ACE2 (focused refinement of RBD and ACE2)
Method: single particle / : Zhu X, Saville JW, Mannar D, Berezuk AM, Cholak S, Tuttle KS, Vahdatihassani F, Subramaniam S

PDB-8dm5: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with human ACE2
Method: single particle / : Zhu X, Saville JW, Mannar D, Berezuk AM, Cholak S, Tuttle KS, Vahdatihassani F, Subramaniam S

PDB-8dm6: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with human ACE2 (focused refinement of RBD and ACE2)
Method: single particle / : Zhu X, Saville JW, Mannar D, Berezuk AM, Cholak S, Tuttle KS, Vahdatihassani F, Subramaniam S

PDB-8dm7: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with mouse ACE2
Method: single particle / : Zhu X, Saville JW, Mannar D, Berezuk AM, Cholak S, Tuttle KS, Vahdatihassani F, Subramaniam S
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
