-Search query
-Search result
Showing 1 - 50 of 10,766 items for (author: hang & j)
EMDB-44255: 
Cryo-EM structure of the mouse TRPM8 channel in the ligand-free desensitized state
Method: single particle / : Yin Y, Park CG, Zhang F, Fedor J, Feng S, Suo Y, Im W, Lee SY

EMDB-44256: 
Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist TC-I 2014
Method: single particle / : Yin Y, Park CG, Zhang F, Fedor J, Feng S, Suo Y, Im W, Lee SY

EMDB-44257: 
Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist AMG2850
Method: single particle / : Yin Y, Park CG, Zhang F, Fedor J, Feng S, Suo Y, Im W, Lee SY
EMDB-44258: 
Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist AMTB
Method: single particle / : Yin Y, Park CG, Zhang F, Fedor J, Feng S, Suo Y, Im W, Lee SY
EMDB-44259: 
Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist TC-I 2014 and the cooling agonist C3
Method: single particle / : Yin Y, Park CG, Zhang F, Fedor J, Feng S, Suo Y, Im W, Lee SY
EMDB-44260: 
Cryo-EM structure of the avian great tit TRPM8 channel in complex with the antagonist TC-I 2014
Method: single particle / : Yin Y, Park CG, Zhang F, Fedor J, Feng S, Suo Y, Im W, Lee SY

EMDB-44261: 
Cryo-EM structure of the mouse TRPM8 channel in complex with PI(4,5)P2 and Ca2+
Method: single particle / : Yin Y, Park CG, Zhang F, Fedor J, Feng S, Suo Y, Im W, Lee SY
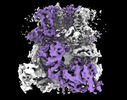
EMDB-44262: 
Cryo-EM structure of the mouse TRPM8 channel in complex with Ca2+ in the absence of PI(4,5)P2
Method: single particle / : Yin Y, Park CG, Zhang F, Fedor J, Feng S, Suo Y, Im W, Lee SY
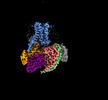
EMDB-60628: 
Carazolol-activated human beta3 adrenergic receptor
Method: single particle / : Zheng S, Zhang S, Dai S, Chen K, Gao K, Lin B, Liu X
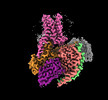
EMDB-60629: 
Epinephrine-activated human beta3 adrenergic receptor
Method: single particle / : Zheng S, Zhang S, Dai S, Chen K, Gao K, Lin B, Liu X

EMDB-60100: 
SARS-CoV-2 spike trimer (6P) in complex with three R1-26 Fabs
Method: single particle / : Yan Q, Gao X, Liu B, Hou R, He P, Li Z, Chen Q, Wang J, He J, Chen L, Zhao J, Xiong X

EMDB-60101: 
SARS-CoV-2 spike trimer (6P) in complex with R1-26 Fab, head-to-head aggregate
Method: single particle / : Yan Q, Gao X, Liu B, Hou R, He P, Li Z, Chen Q, Wang J, He J, Chen L, Zhao J, Xiong X

EMDB-60102: 
SARS-CoV-2 spike trimer (6P) in complex with R1-26 Fab, focused refinement of RBD-Fab region
Method: single particle / : Yan Q, Gao X, Liu B, Hou R, He P, Li Z, Chen Q, Wang J, He J, Chen L, Zhao J, Xiong X

EMDB-60103: 
SARS-CoV-2 spike trimer (6P) in complex with two H18 Fabs
Method: single particle / : Yan Q, Gao X, Liu B, Hou R, He P, Li Z, Chen Q, Wang J, He J, Chen L, Zhao J, Xiong X

EMDB-60104: 
SARS-CoV-2 spike trimer (6P) in complex with three H18 Fabs
Method: single particle / : Yan Q, Gao X, Liu B, Hou R, He P, Li Z, Chen Q, Wang J, He J, Chen L, Zhao J, Xiong X

EMDB-60105: 
SARS-CoV-2 spike trimer (6P) in complex with three H18 Fabs, head-to-head aggregate (C1 symmetry)
Method: single particle / : Yan Q, Gao X, Liu B, Hou R, He P, Li Z, Chen Q, Wang J, He J, Chen L, Zhao J, Xiong X

EMDB-60106: 
SARS-CoV-2 spike trimer (6P) in complex with three H18 Fabs, head-to-head aggregate (C3 symmetry)
Method: single particle / : Yan Q, Gao X, Liu B, Hou R, He P, Li Z, Chen Q, Wang J, He J, Chen L, Zhao J, Xiong X

EMDB-60107: 
SARS-CoV-2 spike trimer (6P) in complex with two H18 and two R1-32 Fabs
Method: single particle / : Yan Q, Gao X, Liu B, Hou R, He P, Li Z, Chen Q, Wang J, He J, Chen L, Zhao J, Xiong X

EMDB-60108: 
SARS-CoV-2 spike trimer (6P) in complex with three H18 and three R1-32 Fabs
Method: single particle / : Yan Q, Gao X, Liu B, Hou R, He P, Li Z, Chen Q, Wang J, He J, Chen L, Zhao J, Xiong X

EMDB-60109: 
SARS-CoV-2 spike trimer (6P) in complex with three H18 and three R1-32 Fabs (one RBD rotated)
Method: single particle / : Yan Q, Gao X, Liu B, Hou R, He P, Li Z, Chen Q, Wang J, He J, Chen L, Zhao J, Xiong X

EMDB-60110: 
SARS-CoV-2 S1 in complex with H18 and R1-32 Fab
Method: single particle / : Yan Q, Gao X, Liu B, Hou R, He P, Li Z, Chen Q, Wang J, He J, Chen L, Zhao J, Xiong X

EMDB-60111: 
Dimer of SARS-CoV-2 S1 in complex with H18 and R1-32 Fabs
Method: single particle / : Yan Q, Gao X, Liu B, Hou R, He P, Li Z, Chen Q, Wang J, He J, Chen L, Zhao J, Xiong X

EMDB-39212: 
Cryo-EM structure of Dragon Grouper nervous necrosis virus-like particle at pH8.0 (3.23A)
Method: single particle / : Wang CH, Chang WH

EMDB-39213: 
Cryo-EM structure of Dragon Grouper nervous necrosis virus-like particle at pH6.5 (2.82A)
Method: single particle / : Wang CH, Chang WH

EMDB-39214: 
Cryo-EM structure of Dragon Grouper nervous necrosis virus-like particle at pH5.0 (3.52A)
Method: single particle / : Wang CH, Chang WH

EMDB-39215: 
Cryo-EM structure of Dragon Grouper nervous necrosis virion at pH6.5 (3.12A)
Method: single particle / : Wang CH, Chang WH

EMDB-39217: 
Cryo-EM structure of Dragon Grouper nervous necrosis virion at pH5.0 (4.36A)
Method: single particle / : Wang CH, Chang WH

PDB-8yf6: 
Cryo-EM structure of Dragon Grouper nervous necrosis virus-like particle at pH8.0 (3.23A)
Method: single particle / : Wang CH, Chang WH

PDB-8yf7: 
Cryo-EM structure of Dragon Grouper nervous necrosis virus-like particle at pH6.5 (2.82A)
Method: single particle / : Wang CH, Chang WH

PDB-8yf8: 
Cryo-EM structure of Dragon Grouper nervous necrosis virus-like particle at pH5.0 (3.52A)
Method: single particle / : Wang CH, Chang WH

PDB-8yf9: 
Cryo-EM structure of Dragon Grouper nervous necrosis virion at pH6.5 (3.12A)
Method: single particle / : Wang CH, Chang WH

EMDB-41770: 
Apo form of human ATE1
Method: single particle / : Huang W, Zhang Y, Taylor DJ

EMDB-42071: 
human ATE1 in complex with Arg-tRNA and a peptide substrate
Method: single particle / : Huang W, Zhang Y, Taylor DJ

PDB-8uau: 
human ATE1 in complex with Arg-tRNA and a peptide substrate
Method: single particle / : Huang W, Zhang Y, Taylor DJ

EMDB-38300: 
Cryo-EM structure of partial dimeric WDR11-FAM91A1 complex
Method: single particle / : Jia GW, Deng QH, Su ZM, Jia D

EMDB-39863: 
Cryo-EM structure of dimeric WDR11-FAM91A1 complex
Method: single particle / : Jia GW, Deng QH, Su ZM, Jia D

EMDB-39943: 
Cryo-EM structure of dimeric WDR11-FAM91A1 complex Body2
Method: single particle / : Jia GW, Deng QH, Su ZM, Jia D

EMDB-39947: 
Cryo-EM structure of dimeric WDR11-FAM91A1 complex Body1
Method: single particle / : Jia GW, Deng QH, Su ZM, Jia D

EMDB-39949: 
Cryo-EM structure of WDR11-dm-FAM91A1 complex
Method: single particle / : Jia GW, Deng QH, Su ZM, Jia D

PDB-8xfb: 
Cryo-EM structure of partial dimeric WDR11-FAM91A1 complex
Method: single particle / : Jia GW, Deng QH, Su ZM, Jia D

PDB-8z9m: 
Cryo-EM structure of dimeric WDR11-FAM91A1 complex
Method: single particle / : Jia GW, Deng QH, Su ZM, Jia D

EMDB-41854: 
Structure of Human Mitochondrial Chaperonin V72I Mutant
Method: single particle / : Chen L, Wang J

PDB-8u39: 
Structure of Human Mitochondrial Chaperonin V72I mutant
Method: single particle / : Chen L, Wang J
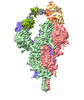
EMDB-37139: 
Structure of SARS-CoV Spike protein complexed with antibody PW5-5
Method: single particle / : Sun L, Mao Q, Wang Y
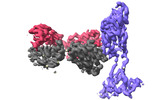
EMDB-37143: 
The local refined map of SARS-CoV-2 XBB Variant Spike protein complexed with antibody PW5-535
Method: single particle / : Sun L, Mao Q, Wang Y
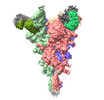
EMDB-37144: 
Trimer state of SARS-CoV Spike protein complexed with antibody PW5-535
Method: single particle / : Sun L, Mao Q, Wang Y

EMDB-37145: 
The local refined map of SARS-CoV Spike protein complexed with antibody PW5-5
Method: single particle / : Sun L, Mao Q, Wang Y
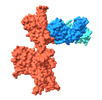
EMDB-37160: 
Monomer state of SARS-CoV-2 XBB Variant Spike protein trimer complexed with antibody PW5-5
Method: single particle / : Sun L, Mao Q, Wang Y
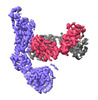
EMDB-37161: 
Monomer state of SARS-CoV Spike protein complexed with antibody PW5-535
Method: single particle / : Sun L, Mao Q, Wang Y
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
