-Search query
-Search result
Showing all 35 items for (author: guenther & s)

EMDB-13867: 
Structure of the bacterial type VI secretion system effector RhsA.
Method: single particle / : Guenther P, Quentin D, Ahmad S, Sachar K, Gatsogiannis C, Whitney JC, Raunser S

PDB-7q97: 
Structure of the bacterial type VI secretion system effector RhsA.
Method: single particle / : Guenther P, Quentin D, Ahmad S, Sachar K, Gatsogiannis C, Whitney JC, Raunser S

EMDB-13843: 
Structure of VgrG1 from Pseudomonas protegens.
Method: single particle / : Guenther P, Quentin D, Ahmad S, Sachar K, Gatsogiannis C, Whitney JC, Raunser S

PDB-7q5p: 
Structure of VgrG1 from Pseudomonas protegens.
Method: single particle / : Guenther P, Quentin D, Ahmad S, Sachar K, Gatsogiannis C, Whitney JC, Raunser S

PDB-6y6k: 
Cryo-EM structure of a Phenuiviridae L protein
Method: single particle / : Vogel D, Thorkelsson SR, Quemin E, Meier K, Kouba T, Gogrefe N, Busch C, Reindl S, Guenther S, Cusack S, Gruenewald K, Rosenthal M

EMDB-9204: 
Cucumber Leaf Spot Virus
Method: single particle / : Sherman MB, Smith TJ

EMDB-9205: 
Red Clover Necrotic Mosaic Virus
Method: single particle / : Sherman MB, Smith TJ

PDB-6mrl: 
Cucumber Leaf Spot Virus
Method: single particle / : Sherman MB, Smith TJ

PDB-6mrm: 
Red Clover Necrotic Mosaic Virus
Method: single particle / : Sherman MB, Smith TJ

EMDB-0535: 
Cryo-EM Reconstruction of Protease-Activateable Adeno-Associated Virus 9 (AAV9-L001)
Method: single particle / : Bennett AB, Agbandje-Mckenna M

PDB-6nxe: 
Cryo-EM Reconstruction of Protease-Activateable Adeno-Associated Virus 9 (AAV9-L001)
Method: single particle / : Bennett AB, Agbandje-Mckenna M

PDB-6hij: 
Cryo-EM structure of the human ABCG2-MZ29-Fab complex with cholesterol and PE lipids docked
Method: single particle / : Jackson SM, Manolaridis I, Kowal J, Zechner M, Taylor NMI, Bause M, Bauer S, Bartholomaeus R, Stahlberg H, Bernhardt G, Koenig B, Buschauer A, Altmann KH, Locher KP

EMDB-7466: 
Segment NFGTFS, with familial mutation A315T and phosphorylated threonine, from the low complexity domain of TDP-43, residues 312-317
Method: electron crystallography / : Guenther EL, Cao Q, Boyer DR, Sawaya MR, Eisenberg DS

EMDB-7467: 
SWGMMGMLASQ segment from the low complexity domain of TDP-43, residues 333-343
Method: electron crystallography / : Guenther EL, Rodriguez JA

EMDB-8857: 
MicroED structure of the segment, NFGEFS, from the A315E familial variant of the low complexity domain of TDP-43, residues 312-317
Method: electron crystallography / : Guenther EL, Sawaya MR

PDB-5wkb: 
MicroED structure of the segment, NFGEFS, from the A315E familial variant of the low complexity domain of TDP-43, residues 312-317
Method: electron crystallography / : Guenther EL, Sawaya MR, Cascio D, Eisenberg DS

PDB-6cf4: 
Segment NFGTFS, with familial mutation A315T and phosphorylated threonine, from the low complexity domain of TDP-43, residues 312-317
Method: electron crystallography / : Guenther EL, Cao Q, Boyer DR, Sawaya MR, Eisenberg DS

PDB-6cfh: 
SWGMMGMLASQ segment from the low complexity domain of TDP-43
Method: electron crystallography / : Guenther EL, Rodriguez JA, Sawaya MR, Eisenberg DS

EMDB-3953: 
Structure of inhibitor-bound ABCG2
Method: single particle / : Jackson SM, Manolaridis I, Kowal J, Zechner M, Altmann KH, Locher KP

EMDB-4246: 
Structure of inhibitor-bound ABCG2
Method: single particle / : Jackson SM, Manolaridis I, Kowal J, Zechner M, Altmann KH, Locher KP

EMDB-4256: 
Structure of an inhibitor-bound ABC transporter
Method: single particle / : Jackson SM, Manolaridis I, Kowal J, Zechner M, Taylor NMI, Bause M, Bauer S, Bartholomaeus R, Stahlberg H, Bernhardt G, Koenig B, Buschauer A, Altmann KH, Locher KP

PDB-6eti: 
Structure of inhibitor-bound ABCG2
Method: single particle / : Jackson SM, Manolaridis I, Kowal J, Zechner M, Altmann KH, Locher KP

PDB-6feq: 
Structure of inhibitor-bound ABCG2
Method: single particle / : Jackson SM, Manolaridis I, Kowal J, Zechner M, Altmann KH, Locher KP

PDB-6ffc: 
Structure of an inhibitor-bound ABC transporter
Method: single particle / : Jackson SM, Manolaridis I, Kowal J, Zechner M, Taylor NMI, Bause M, Bauer S, Bartholomaeus R, Stahlberg H, Bernhardt G, Koenig B, Buschauer A, Altmann KH, Locher KP

EMDB-8781: 
CryoEM structure of the segment, DLIIKGISVHI, assembled into a triple-helical fibril
Method: helical / : Guenther EL, Ge P

PDB-5w7v: 
CryoEM structure of the segment, DLIIKGISVHI, assembled into a triple-helical fibril
Method: helical / : Guenther EL, Ge P, Eisenberg DS

EMDB-8765: 
MicroED structure of the segment, DLIIKGISVHI, from the RRM2 of TDP-43, residues 247-257
Method: electron crystallography / : Guenther EL, Sawaya MR

PDB-5w52: 
MicroED structure of the segment, DLIIKGISVHI, from the RRM2 of TDP-43, residues 247-257
Method: electron crystallography / : Guenther EL, Sawaya MR, Cascio D, Eisenberg DS

EMDB-3001: 
MicroED structure of the segment, GVVHGVTTVA, from the A53T familial mutant of Parkinson's disease protein, alpha-synuclein, residues 47-56
Method: electron crystallography / : Rodriguez JA, Ivanova M, Sawaya MR, Cascio D, Reyes F, Shi D, Johnson L, Guenther E, Sangwan S, Hattne J, Nannenga B, Gonen T, Eisenberg D
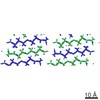
EMDB-3028: 
MicroED structure of the toxic core segment, GAVVTGVTAVA, from Parkinson's disease protein, alpha-synuclein, residues 69-78.
Method: electron crystallography / : Rodriguez JA, Ivanova M, Sawaya MR, Cascio D, Reyes F, Shi D, Johnson L, Guenther E, Zhang M, Jiang L, Arbing MA, Sangwan S, Hattne J, Whitelegge J, Brewster A, Messerschmidt M, Boutet S, Sauter NK, Nannenga B, Gonen T, Eisenberg D

PDB-4znn: 
MicroED structure of the segment, GVVHGVTTVA, from the A53T familial mutant of Parkinson's disease protein, alpha-synuclein residues 47-56
Method: electron crystallography / : Rodriguez JA, Ivanova M, Sawaya MR, Cascio D, Reyes F, Shi D, Johnson L, Guenther E, Sangwan S, Hattne J, Nannenga B, Brewster AS, Messerschmidt M, Boutet S, Sauter NK, Gonen T, Eisenberg DS

PDB-4ril: 
Structure of the amyloid forming segment, GAVVTGVTAVA, from the NAC domain of Parkinson's disease protein alpha-synuclein, residues 68-78, determined by electron diffraction
Method: electron crystallography / : Rodriguez JA, Ivanova M, Sawaya MR, Cascio D, Reyes F, Shi D, Johnson L, Guenther E, Sangwan S, Hattne J, Nannenga B, Brewster AS, Messerschmidt M, Boutet S, Sauter NK, Gonen T, Eisenberg DS

EMDB-5369: 
Structural transitions in RCNMV revealing potential mechanism of RNA release (native map)
Method: single particle / : Sherman MB, Guenther RH, Tama F, Sit TL, Brooks CL, Mikhailov AM, Orlova EV, Baker TS, Lommel SA

EMDB-5372: 
Structural transitions in RCNMV revealing potential mechanism of RNA release (EGTA map)
Method: single particle / : Sherman MB, Guenther RH, Tama F, Sit TL, Brooks CL, Mikhailov AM, Orlova EV, Baker TS, Lommel SA

EMDB-5373: 
Structural transitions in RCNMV revealing potential mechanism of RNA release (EDTA map)
Method: single particle / : Sherman MB, Guenther RH, Tama F, Sit TL, Brooks CL, Mikhailov AM, Orlova EV, Baker TS, Lommel SA
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
